Going the Distance: FUS Travels to Terminals, Drops Off RNA
Quick Links
Following their transcription from the genome, RNA messages exit the nucleus and enter the cytoplasm, sometimes traveling vast distances before they are finally translated into protein. RNA-binding proteins are helping hands on this journey from start to finish. At the 5th RNA Metabolism in Neurological Disease Conference, held November 1‒2 in San Diego, scientists shared news about FUS. This RNA-binding protein not only accompanies RNA cargo on its trip to distant axons for translation, it also requires its own chaperones to avoid getting sidetracked into globs of liquid droplets called ribonucleoprotein granules along the way. This careful shepherding gets derailed when FUS carries mutations known to cause ALS/FTD. Finally, researchers raised the possibility that dalliances with chaperones and specialized ribosomes in the neuronal synapse might push FUS into releasing its precious RNA cargo for axonal translation.
- FUS mutants trigger a cell stress response, shut down translation in axons.
- The nuclear transportin TNPO1 chaperones FUS in cytoplasm, preventing aggregation.
- Specialized ribosomes interact with FUS; translate specific mRNAs.
Sandrine Da Cruz of the University of California, San Diego, showed recently published data suggesting that cellular stress derails axonal translation when FUS is mutated (Lopez-Erauskin et al., 2018). The researchers used a bacterial artificial chromosome to express human FUS under control of the mouse promoter. Mice lacking the FUS gene die before birth, but live when expressing the human gene. By all accounts, human wild-type FUS functionally replaced the mouse gene, but mice that expressed either of two ALS-linked FUS mutants developed an ALS-like disease marked by a weakening grip and loss of a third of their neuromuscular junctions by 18 months of age. By two years, the mice started slipping on memory tests, and neurons in their hippocampi had fewer synapses.
Da Cruz reported that in both wild-type and mutant mice, the majority of FUS protein was in the nucleus; she saw no cytoplasmic aggregates. What’s more, Da Cruz saw no obvious changes in expression or splicing of genes normally affected by loss of FUS expression. This means that FUS mutations cause neurodegeneration in ways other than altering FUS function in the nucleus.
What could it be? Da Cruz saw downregulation of genes encoding the translational machinery, including ribosomal proteins, in the spinal cords of mice expressing mutant FUS. Expression of genes encoding key synaptic proteins also took a plunge. The spinal cord was marked by increased phosphorylation of eIF2α, which shuts off translation in response to cellular stress. The twist: This stress response and translational shutdown happened only in axons. By labeling newly synthesized proteins with puromycin, Da Cruz found blocked protein synthesis in axons, not cell bodies, of neurons expressing mutant FUS. This was true in cultured hippocampal neurons and in sciatic nerves of mice. Furthermore, a higher proportion of FUS accumulated in axons of mutant FUS mice compared with wild-type FUS mice.
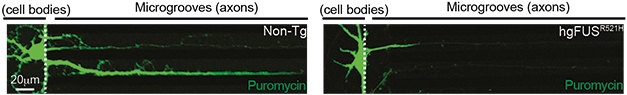
Axons Stopped in Translation. Newly synthesized proteins (green, puromycin) populate both cell bodies and axons of hippocampal neurons from non-transgenic mice (left). Only cell bodies have substantial puromycin in FUS mutant mice (right) [Courtesy of Lopez-Erauskin et al., Neuron, 2018.]
Da Cruz believes mutant FUS somehow triggers the integrated stress response in axons, halting translation locally. Some quibbled that the dearth of newly synthesized proteins in axons could be explained by a deficit in axonal transport, or by the general degeneration of axons. Da Cruz replied that she sees this deficit in axonal, not somal, translation almost immediately after injecting puromycin, suggesting the defect was localized and specific. Exactly how mutant FUS incites this response, and what physiological role FUS normally plays in axonal transport and translation, remains unclear. Fielding more questions after her talk, Da Cruz said her group searched for signs of stress granules or FUS aggregates in axons, but found none.
Dorothee Dormann of Ludwig-Maximilians-University, Munich, described a mechanism that could sidetrack FUS in the cytoplasm, possibly preventing the RNA-binding protein from delivering its cargo to sites of translation. As described in Alzforum’s recent coverage of Dormann’s paper, under normal conditions, FUS interacts with the nuclear import receptor TNPO1 (Apr 2018 news on Hofweber et al., 2018). This protein chaperones FUS in the cytoplasm and keeps it from coalescing into RNA granules. In this way, TNPO1 allows FUS to travel, unfettered, to sites of translation. ALS/FTD mutations hinder this interaction, rendering FUS prone to sequestration.
Dormann found that besides binding to FUS’s nuclear localization sequence, TNPO1 can also latch onto its RNA-binding domain, competing with RNA for attachment to FUS. In San Diego, Dormann said that she spotted TNPO1 lingering in RNA granules way out in the neurites of primary rat cortical neurons. She thinks TNPO1 might outcompete FUS-bound RNA there, thus releasing transcripts for local translation.
Roy Parker of the University of Colorado in Boulder told Alzforum that Dormann’s findings underscore the importance of maintaining a dynamic environment in the cytoplasm. “Just as RNA binding proteins like FUS and TDP-43 prevent RNA entanglement, it seems that chaperones like TNPO1 prevent these RNA-binding proteins from becoming ensnared in granules,” he said.
Peter St George-Hyslop’s data meshed with Dormann’s. His lab at the University of Toronto focused on the dynamics of FUS phase separation in axonal terminals. He found that both arginine hypomethylation and ALS/FTD mutations promote FUS phase separation and keep FUS enmeshed in granules. Similar to Dormann, he reported that TNPO1 counters this gelling tendency. If FUS remains stuck in granules, local translation at axonal terminals cannot happen (Apr 2018 news on Qamar et al., 2018). Likening granules to Borg assimilation in "Star Trek," St George-Hyslop said that occasionally, FUS granules merge, creating an ever-larger trap for FUS, RNA, and possibly translational machinery.
The translation machinery itself drew attention at this workshop. It includes a distinctive palette of proteins depending on where in the cell, and under what conditions, translation takes place, according to Maria Barna of Stanford University in Palo Alto, California. Barna described a startlingly diverse world of specialized ribosomes. Cast as blobs floating in a vast cytoplasmic sea, ribosomes have long been considered boring workhorses that translate any old transcript they come across. Barna’s work casts ribosomes as sites of exquisite specificity.
Using triple quadrupole mass spectrometry, Barna found that a fraction of the 80 core ribosomal proteins only appear in a subset of so-called specialized ribosomes. Strikingly, Barna reported that specialized ribosomes translated mRNAs involved in distinctive processes or pathways. For example, ribosomes containing the protein Rp10a tended to translate mRNAs involved in cell growth, but avoided mRNAs linked to opposing processes such as cell stress or apoptosis. Comparing specialized ribosomes with a bacterial operon system, Barna reported that certain ribosomes were responsible for translating proteins involved in entire pathways, such as vitamin B12 biosynthesis (Shi et al., 2017). The cadre of specialized ribosomes also shifted during cellular differentiation from stem cells to other cell types, according to preliminary data from her lab.
Barna also sees this specificity in the ribosomal interactome. She identified more than 200 ribosome-associated proteins, aka RAPs. They differed based on cell type and intracellular location. Barna has yet to investigate the landscape of specialized ribosomes or RAPs within neurons, but proposed that they could form the basis for specific translation in the axon or synapse. One tidbit she does know already: the ALS/FTD related proteins FUS, VCP, and FMRP1 are among the proteins that interacted with subsets of ribosomes (Simsek et al., 2017).
Da Cruz wondered whether these interactions might explain the specialized delivery of mRNAs by FUS or other RNA binding proteins out to the synapses. Dovetailing with Dormann’s idea that TNPO1-FUS interactions at the synapse help FUS shed its mRNA cargo there, perhaps associations with specialized ribosomes come into play as well, she speculated.
Benjamin Wolozin of Boston University told Alzforum that Barna’s findings about ribosomal complexity are an important factor the neurodegenerative disease field has yet to address. Specialized ribosomes could play a role in the selective vulnerability of neurons to disease.—Jessica Shugart
References
News Citations
Paper Citations
- López-Erauskin J, Tadokoro T, Baughn MW, Myers B, McAlonis-Downes M, Chillon-Marinas C, Asiaban JN, Artates J, Bui AT, Vetto AP, Lee SK, Le AV, Sun Y, Jambeau M, Boubaker J, Swing D, Qiu J, Hicks GG, Ouyang Z, Fu XD, Tessarollo L, Ling SC, Parone PA, Shaw CE, Marsala M, Lagier-Tourenne C, Cleveland DW, Da Cruz S. ALS/FTD-Linked Mutation in FUS Suppresses Intra-axonal Protein Synthesis and Drives Disease Without Nuclear Loss-of-Function of FUS. Neuron. 2018 Nov 21;100(4):816-830.e7. Epub 2018 Oct 18 PubMed.
- Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T, Dormann D. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell. 2018 Apr 19;173(3):706-719.e13. PubMed.
- Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Ströhl F, Meadows W, Ferry R, Dardov VJ, Tartaglia GG, Farrer LA, Kaminski Schierle GS, Kaminski CF, Holt CE, Fraser PE, Schmitt-Ulms G, Klenerman D, Knowles T, Vendruscolo M, St George-Hyslop P. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell. 2018 Apr 19;173(3):720-734.e15. PubMed.
- Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, Barna M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol Cell. 2017 Jul 6;67(1):71-83.e7. Epub 2017 Jun 15 PubMed.
- Simsek D, Tiu GC, Flynn RA, Byeon GW, Leppek K, Xu AF, Chang HY, Barna M. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell. 2017 Jun 1;169(6):1051-1065.e18. PubMed.
Further Reading
No Available Further Reading
Primary Papers
- López-Erauskin J, Tadokoro T, Baughn MW, Myers B, McAlonis-Downes M, Chillon-Marinas C, Asiaban JN, Artates J, Bui AT, Vetto AP, Lee SK, Le AV, Sun Y, Jambeau M, Boubaker J, Swing D, Qiu J, Hicks GG, Ouyang Z, Fu XD, Tessarollo L, Ling SC, Parone PA, Shaw CE, Marsala M, Lagier-Tourenne C, Cleveland DW, Da Cruz S. ALS/FTD-Linked Mutation in FUS Suppresses Intra-axonal Protein Synthesis and Drives Disease Without Nuclear Loss-of-Function of FUS. Neuron. 2018 Nov 21;100(4):816-830.e7. Epub 2018 Oct 18 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.