CRISPR Screen Pulls Down Fresh Targets for C9ORF72 ALS
Quick Links
Protein products of an expanded repeat sequence in the C9ORF72 gene harm neurons in myriad ways, ultimately leading to amyotrophic lateral sclerosis (ALS) and/or frontotemporal dementia (FTD). In a March 5 study in Nature Genetics, researchers led by Aaron Gitler and Michael Bassik at Stanford University deployed CRISPR gene editing to screen for modifiers of this toxicity in human cells and in mouse neurons. They pulled out hundreds of genes that either helped or hindered C9’s damage from multiple pathways, including nucleocytoplasmic transport, RNA processing, and endoplasmic reticulum (ER) stress. Knocking out one of the most potent modifiers—a gene called TMX2—dampened harmful cell stress signals and boosted survival of neurons from patients with C9-ALS.
- A CRISPR screen in human cells and in mouse neurons revealed modifiers of toxicity inflicted by dipeptide repeats translated from C9ORF72.
- Top hits pointed toward nucleocytoplasmic transport, RNA processing, and ER stress.
- Deleting the TMX2 gene in mouse neurons or patient-derived neurons promoted healthy responses to stress.
“[The study] truly shows that CRISPR/Cas9 is not only a tool for therapeutic genome editing but, compared to other genome-editing approaches, has the unique capacity for genome-wide screens and biological discovery,” commented David Schaffer of University of California, Berkeley.
Robert Baloh of Cedars Sinai Medical Center in Los Angeles commended the authors, saying that the powerful screens identified unanticipated modifiers of C9ORF72 toxicity. “The beauty of their approach is that it immediately provides a target for drug screening, and a therapeutic avenue toward the clinic,” he added.
Aggregates of dipeptide repeats (DPRs) translated from C9ORF72 repeat expansions are known to disrupt multiple cellular functions. Five types of DPR are known, of which two arginine-containing varieties—glycine-arginine (GR) and proline-arginine (PR)—are the most toxic. Researchers know that the peptides link up with proteins that contain low-complexity domains, including RNA-binding proteins, parts of the ribosome and other translation factors, and the nuclear pore complex. In this way, the DPRs derail multiple cellular processes (Aug 2014 news; Dec 2014 news; Aug 2015 news; Oct 2016 news). The DPRs’ promiscuity offers rich opportunities to try to stem the damage they cause. To date, researchers have performed genome-wide screens for modifiers of DPR toxicity in model organisms, including yeast and flies, but screens in mammalian systems have awaited technical advances in gene editing (Jovičić et al., 2015; Boeynaems et al., 2016).
Enter CRISPR. Co-first authors Nicholas Kramer and Michael Haney used the gene-editing tool to perform the first-ever screen for genetic modifiers of DPR toxicity in mammalian cells. They used a human myelogenous leukemia cell line, K562, because the cells grow rapidly and have proven their worth in other genome-wide screens. The cells stably express the Cas9 nuclease, which targets single-guide RNAs (sgRNAs) designed to adhere to target genes. For the DPR screen, the researchers transduced the cells with a library of 10 sgRNAs per each of 20,500 genes, as well as 10,000 negative control sgRNAs. They then treated the cells with synthetic polymers of PR20 or GR20 at doses known to kill half of the cells, and later performed deep sequencing on the remaining cells to determine which CRISPR knockout clones were enriched (i.e., protected) versus depleted (i.e., sensitized) to the DPR assault.
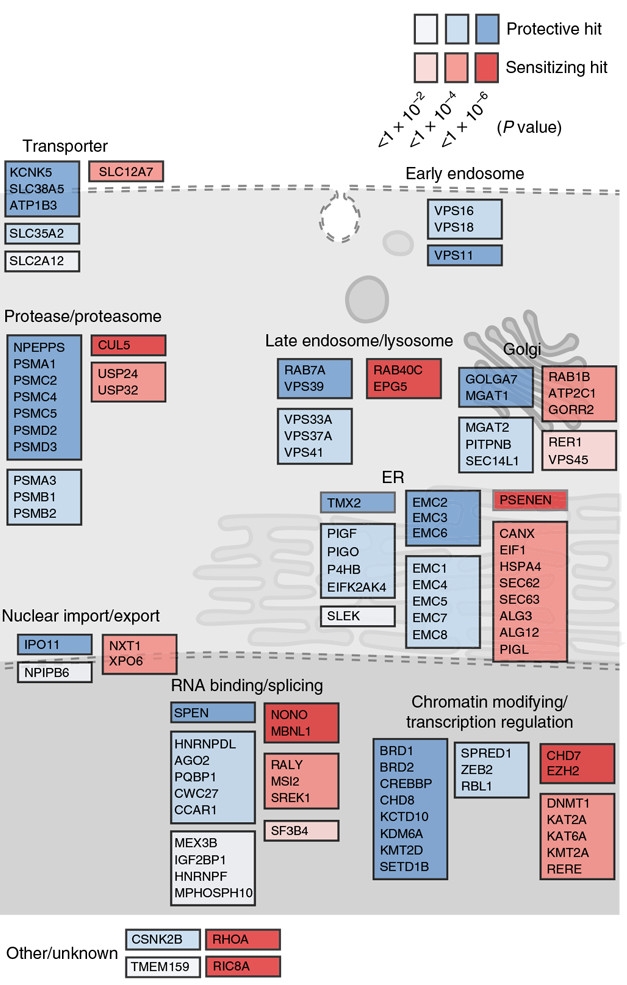
CRISP New Targets? Top modifiers of DPR toxicity, arranged by estimated cellular function. Protective hits (blue) boosted survival when knocked out, while sensitizing hits (red) killed cells. [Courtesy of Kramer et al., Nature Genetics, 2018.]
The scientists identified 215 and 387 genetic modifiers of PR20 and GR20 toxicity, respectively, many of which overlapped. Among the top hits were some of the same nuclear transport genes identified in yeast and fly screens, suggesting common mechanisms of toxicity across organisms. There were also brand-new modifiers, including genes encoding RNA-binding proteins, splicing machinery, chromatin modifiers, and transcriptional regulators. Interestingly, many genes encoding proteins that reside in the ER popped up, including those involved with the ER-associated degradation pathway that switches on in response to cell stress.
Would these genes also modify C9ORF72 toxicity in neurons? To find out, the researchers ran toxicity screens in mouse primary cortical neurons derived from animals expressing Cas9. This time, they delivered the toxic DPRs via two different routes: either by adding synthetic PR20 to the culture media, as they had done in the human screen, or by using a lentivirus to express PR50 within the cells. That way they could distinguish between genes that played a role in uptake and intracellular transport of the peptides, and those that mediated toxic effects after cellular entry. They transduced the cells with a library of sgRNAs designed to target the top 200 hits from the human genome-wide screen.
They identified 13 modifiers from the synthetic PR20 screen, and 17 from the viral-expression screen. Only five overlapped, meaning that toxicity modifiers varied significantly depending on the mode of DPR delivery. The researchers reasoned that modifiers identified only in the synthetic PR20 screen might be relevant if DPRs spread from cell to cell within the brain. The endolysosomal trafficking gene, RAB7A, emerged as a top hit from that screen. Investigating this further, the researchers confirmed that knocking out RAB7A in HeLa cells prevented synthetic PR20 polymers from reaching the nucleus, where they are known to do damage. Instead, the externally administered dipeptides remained trapped in early endosomal compartments.
The top hits in the lentiviral PR50 screen were primarily nuclear and ER-resident proteins. TMX2 was a top modifier, as neurons lacking the gene had a clear survival advantage. This gene also modified toxicity in the human screen. Little is known about the gene, except that it encodes an ER transmembrane thioredoxin protein with uncertain activity. Other members of the TMX family have been implicated in modulating the ER stress response.
Taking a step back, the researchers first assessed whether this stress response in fact switched on when PR50 was expressed in neurons. Indeed, a transcriptional analysis revealed an induction of multiple stress-associated genes in response to PR50 expression, including genes involved in DNA repair, translation, and apoptosis. The same was true in human K562 cells treated with synthetic PR20. The findings pointed to the integrated stress response (ISR), which converges on the phosphorylation of eIF2a and the global inhibition of translation. Indeed, pretreating the K562 cells with ISRIB, an inhibitor of eIF2a phosphorylation, substantially reduced the toxicity inflicted by PR20.
Reasoning that TMX2 could play a role in evoking this harmful stress response, the researchers next sequenced the RNA of the mouse neurons expressing sgRNAs targeting TMX2. Compared with neurons expressing control sgRNAs, the TMX2-knockout neurons responded to PR50 expression by ramping up pro-survival genes and turning down apoptotic ones. In particular, the cells switched on genes involved in the unfolded protein response, a wing of the ER stress response that tends to promote survival. Atf3, a gene already deemed protective in other mouse models of ALS, was among those induced by DPRs in the neurons lacking TMX2. Strikingly, the researchers found that overexpressing Atf3 in mouse neurons protected them from DPR toxicity. Haney told Alzforum that overall, the findings suggest that TMX2 somehow hinders protective responses to DPRs, and promotes destructive ones. However, the details of the relationship between TMX2 and Atf3 remain to be elucidated.
Finally, the researchers sought to extend their findings into patient-derived neurons. They generated induced motor neurons (iMNs) from induced pluripotent stem cells (iPSCs) derived from patients with C9-ALS or healthy controls. Compared with controls, cells derived from C9-ALS patients rapidly succumbed to treatment with glutamate, a standard test of cellular vigor. However, when the researchers knocked down expression of TMX2 using short hairpin RNAs, survival of two different C9-ALS iMN lines in the face of glutamate treatment rose significantly.
“The data in iPSC-derived motor neurons, while modest in effect, is really a key finding in this the paper,” commented Peter Todd of the University of Michigan in Ann Arbor. “It is the only part of the manuscript done at endogenous levels in the setting of the GGGGCC repeat and further supports the argument that DPRs, and PR and GR in particular, are drivers of toxicity in the disorder,” he added.
The researchers interpreted their findings to indicate that TMX2 skews the response to ER stress toward cell death, while removing TMX2 steers the cell toward life-saving maneuvers. They aim to investigate the physiological role of this gene further, perhaps in mouse models of C9ORF72 toxicity, Kramer said.
Todd commented that the paper adds to growing evidence that DPRs stoke the fire of cellular stress. Two recent papers—one from Todd’s group and the other led by Shuying Sun of Johns Hopkins University in Baltimore—reported that the integrated stress response promoted translation of DPRs while suppressing global translation (Jan 2018 news). This is because the DPRs are translated via a noncanonical mechanism called repeat-associated non-AUG (RAN) translation. Sun noted that although the DPRs in Gitler’s screen were not expressed via RAN, together they pointed to the existence of a toxic feed-forward loop, in which cellular stress in response to DPRs also fuels their enhanced production. However, Sun cautioned that because RAN translation was not addressed in the screen, it is possible that targeting some modifiers, such as TMX2, might elevate such translation in its physiological context.
Todd added that while these details need to be ironed out, the paper supports the idea that targeting the ER stress pathway is a path forward. “Whether a more targeted approach, such as targeting TMX2, or a factor specifically required for RAN translation, or a broad approach such as application of ISRIB to block the whole pathway is more sensible will be a point of discussion in the field going forward,” he wrote.
What about other hits? Kramer and Haney told Alzforum that they hope the screen results will serve as a continuing resource for the ALS community, and that researchers will investigate modifiers that interest them.
Ludo Van Den Bosch of KU Leuven in Belgium agreed. “It is very clear that the information obtained from these genome-wide screens will be an important source of modifiers that can be tested for their effect in other systems, and that could lead to the development of new therapeutic strategies,” he wrote to Alzforum.—Jessica Shugart
References
News Citations
- C9ORF72 Killer Dipeptides Clog the Nucleolus
- Live-Cell Studies Blame Arginine Peptides for C9ORF72’s Crimes
- ALS Gene Repeats Obstruct Traffic Between Nucleus and Cytoplasm
- ALS Research ‘Gels’ as Studies Tie Disparate Genetic Factors Together
- Stressed-Out Cells Translate C9ORF72 Repeats, Unleash Toxic Peptides
Paper Citations
- Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G, Kramer NJ, Gage FH, Van Den Bosch L, Robberecht W, Gitler AD. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015 Sep;18(9):1226-9. PubMed.
- Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovičić A, De Baets G, Scheveneels W, Steyaert J, Cuijt I, Verstrepen KJ, Callaerts P, Rousseau F, Schymkowitz J, Cruts M, Van Broeckhoven C, Van Damme P, Gitler AD, Robberecht W, Van Den Bosch L. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016 Feb 12;6:20877. PubMed.
Further Reading
No Available Further Reading
Primary Papers
- Kramer NJ, Haney MS, Morgens DW, Jovičić A, Couthouis J, Li A, Ousey J, Ma R, Bieri G, Tsui CK, Shi Y, Hertz NT, Tessier-Lavigne M, Ichida JK, Bassik MC, Gitler AD. CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat Genet. 2018 Apr;50(4):603-612. Epub 2018 Mar 5 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Director of Neuromuscular Medicine, Cedars-Sinai Medical Center
This is another great paper by the Gitler and Bassik groups, using the power of genetic screens to identify previously unanticipated modifiers of dipeptide toxicity from C9ORF72 repeat expansion. The beauty of their approach is that it immediately provides a target for drug screening, and a therapeutic avenue toward the clinic. It will be very exciting to see whether manipulating TMX2 can be used for therapeutic intervention in C9ORF72 ALS/FTD in the future.
Neurobiology, KU Leuven & VIB
The group of Aaron Gitler is the first one to successfully perform a genome-wide gene knockout screen using CRISPR–Cas9 in a human cells system to search for suppressors and enhancers of C9ORF72 DPR toxicity.
First of all, this study provides clear proof of principle that such a CRISPR-Cas9 screen in the context of ALS is feasible in human cells, as well as in mouse primary cortical neurons. Moreover, and very importantly, the obtained hits were unique for the DPR-induced toxicity and didn’t overlap with hits of other unrelated toxicity screens.
Second, it is very encouraging that there is considerable overlap between the hits in the PR and GR toxicity screen. This indicates that similar pathways are involved in the induction of toxicity, which is in line with what one could expect.
Third, these CRISPR-Cas9 screens could also provide interesting new insights into the pathways playing a role in the toxicity induced by synthetic arginine-containing DPRs or PR translated from a codon-optimized construct. At present, the screen using the human cell system especially confirms the importance of a number of pathways that have already been linked to DPR-induced toxicity in other systems, which is also very encouraging. These include nucleocytoplasmic transport, endoplasmic reticulum (ER) stress, proteasomal dysfunction, involvement of RNA-processing as well as of chromatin modifications. A very interesting observation is the involvement of RAB7A and the endosomal pathway in the uptake of DPRs. This could have important implications for the cell-to-cell spreading of DPRs.
Finally, it is very interesting that one modifier, TMX2, modulated the ER-stress signature elicited by C9ORF72 DPRs in neurons and also improved survival of human induced motor neurons (iMNs) from patients with C9ORF72 ALS. Moreover, the major importance of ER stress could also be confirmed pharmacologically.
In conclusion, it is very clear that the information obtained from these genome-wide screens will be an important source of modifiers that can be tested for their effect in other systems, and that could lead to the development of new therapeutic strategies. Moreover, there is no doubt that more genome-wide CRISPR-Cas9-related screens will follow in other cell systems and in other toxicity paradigms related to ALS, given the fact that the feasibility and the potential success is convincingly shown by the current study.
Institute for Multigenerational Studies
This work merits attention. It again truly shows that CRISPR/Cas9 is not only a tool for therapeutic genome editing but, compared with other genome-editing approaches, has the unique capacity for genome-wide screens and biological discovery. One can hope that the results of this work point to general mechanisms of ALS pathology that translate to other genotypes and sporadic cases, such that we can begin to consider genome editing or other approaches to develop a broad therapy for this devastating disease.
University of Sheffield
Debate over dipeptide repeats (DPRs) continues, but undoubtedly they are toxic in culture. This important study from Aaron Gitler and Michael Bassik describes the first genome-wide screen for modifiers of DPR toxicity in a mammalian cell line. The genome-wide section of the work utilized K562 cells exposed to exogenous poly proline-arginine (PR) or poly glycine-arginine (GR). Interestingly, there was significant correlation between hits in response to both proteins (but zero overlap with a ricin toxicity screen), suggesting a common pathway for toxicity. This is not a “given” and will help to direct future model design centered on DPRs.
Hits from the K562 screen were validated in a number of ways, including exposing mouse primary cortical neurons to exogenous PR or using a lentivirus to induce endogenous PR expression. Interestingly, the method of PR delivery appeared to have a significant effect—both models validated hits from the original screen but the overlap was relatively small. This is particularly interesting when it is considered that neither system included RAN translation, which is the method by which DPR are produced in patients.
Top hits, which were extensively validated, implicate the ER stress response and, in the context of extracellular PR, the intracellular trafficking of PR. A number of other novel and established biological pathways were highlighted including nucleocytoplasmic transport.
The authors rightly highlight the current lack of supporting evidence of DPR toxicity in postmortem studies but in vitro, the field is making good progress towards therapeutic targets.
University of Michigan
Here are my thoughts on the paper.
Make a Comment
To make a comment you must login or register.