FUS Helps miRNA Silence Genes. For ALS, What Does It Mean?
Quick Links
The more researchers probe and prod the ALS-related protein FUS, the more things they find it does. The latest function, reported in the March 1 Molecular Cell, is gene silencing mediated by micro RNAs. Using mouse and human cellular assays, researchers led by Jiou Wang, Johns Hopkins University, Baltimore, found that FUS binds to the protein Argonaute, to miRNAs, and their mRNA targets, promoting gene silencing. A FUS mutant that causes a severe form of ALS performs less well than wild-type FUS, but whether this contributes to pathogenesis is unknown.
- FUS interacts with gene-silencing components Argonaute 2, miRNAs, and their target mRNAs.
- Optimal miRNA-mediated silencing requires FUS.
- The R495X FUS mutant underperforms wild-type FUS, but the implications for disease are unclear.
“This interesting paper adds a further twist to the ongoing story,” said Yves Goldberg, INSERM-Université Grenoble Alpes in France. “It is not surprising that FUS has a connection with miRNA; what is new is FUS interacting with Argonaute in the cytoplasm and acting directly on translation.”
More than 50 mutations in FUS have been found in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), but how they cause disease remains unclear. FUS wears many hats in the nucleus. It functions in DNA repair, transcription, pre-mRNA splicing, alternative poly-adenylation, RNA transport, and the production of micro RNAs (miRNAs). In neurons, it may also help shuttle and translate mRNA at synapses (for review see Deng et al., 2014).
A first clue that FUS might be busying itself with yet another task came from a screen for proteins that interact with Argonaute 2. AGO2 is a protein at the core of the gene silencing complex miRISC. First authors Tao Zhang and Yen-Ching Wu identified several AGO2 interactors, but FUS caught their eyes because of its robust binding and link to ALS. The researchers used co-immunoprecipitation to confirm the two proteins indeed form a pair in mouse forebrain lysates and embryonic fibroblasts. To map where they bind, the authors expressed a series of truncated FUS and AGO2 proteins in human embryonic kidney cells (HEK293) and checked each construct’s ability to pull down its partner. Binding mapped to the middle domain of AGO2 and the RGG2 domain of FUS.
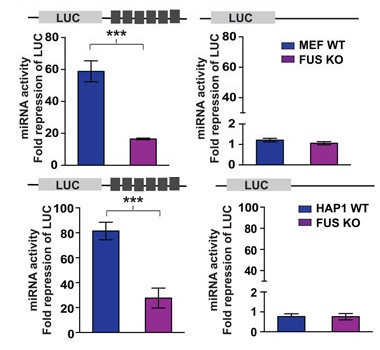
No FUS, No Silence.
Mouse fibroblasts (top) and human HAP1 cells (bottom) expressing wild-type FUS (blue) repress expression of a reporter of miRNA activity three times more than do cells lacking FUS (purple). For a control reporter without silencing sites (right), the presence or absence of FUS makes no difference. [Zhang et al., Mol Cell, 2018]
FUS had been previously reported to help make miRNAs in the nucleus (Morlando et al., 2012; Modigliani et al., 2014). However, the newly found interaction with AGO2 suggests FUS might also help miRNAs degrade their target mRNAs in the cytoplasm. To test this, the researchers used cells that express mRNA encoding the light-generating protein luciferase with a modified 3' UTR that can be silenced by a small interfering RNA (siRNA). The siRNA mimics miRNA activity but bypasses miRNA production. When the authors knocked out FUS from mouse embryonic fibroblasts and a human HAP1 cell line, the cells lit up three times brighter than controls, indicating FUS is needed for full-strength gene silencing (image at left). The researchers then monitored luciferase activity in the presence of R495X, a human ALS FUS mutant lacking part of the RGG2 domain. These cells shined twice as bright as cells expressing wild-type FUS, indicating reduced Argonaute 2 binding renders FUS less able to boost miRNA activity.
The researchers used microarrays to survey mRNA and miRNA levels in cells expressing R495X. Many miRNAs were slightly more abundant in these cells than in wild-type FUS controls, but levels of their target mRNAs were as much as sevenfold greater, confirming that reduced FUS-AGO2 binding disables miRNA activity. Because R495X also misses the FUS nuclear localization signal, Wang acknowledged that change in other FUS functions may contribute to the observed effects.
How does FUS interact with miRNA and mRNA? The researchers focused on a specific miRNA, miR-200c, and one of its targets, ZEB-1. In sum, they found that FUS binds not only to AGO2, but also to miR-200c and ZEB-1, the latter being dependent on miR-200c. FUS and miR-200c occupied neighboring sites on the ZEB-1 3'UTR.
Wang does not know yet how FUS helps degrade mRNA targets, but he has ideas. “FUS may help miRNAs find their transcript targets and bring them together with AGO2,” he said, noting the challenge of doing this in a cell bustling with thousands of mRNAs. FUS could also facilitate AGO2 loading, act as a transient chaperone, stabilize miRNA binding, or form a membrane-less compartment through liquid-liquid phase separation.
Udai Pandey, University of Pittsburgh, finds the study interesting because it reveals how mutations in FUS could have even more widespread consequences than previously thought. This is consistent with the aggressive, early onset ALS caused by several FUS mutations, including R495X. Pandey added that the authors’ finding that a C. elegans homolog of FUS facilitates miRNA silencing indicates the pathway is highly conserved. On the flip side, Goldberg noted that it is harder to explain motor neuron selectivity with a perturbation of this sort.
Given the panoply of FUS functions, what might this new one contribute to ALD/FTD? “Of course the dream would be to identify a single target, but we have all these functions. It is highly frustrating from a medical perspective,” said Goldberg. Wang agreed, but added, “It is important to understand the biology to eventually make corrections.” A deeper basic understanding could also lead to the discovery of biomarkers, he said.
Wang wants to look for similar functions in other disease-related RNA-binding proteins. A candidate is TDP-43, which is also involved in miRNA production. Pandey would like to see whether other FUS mutations, such as P525L, similarly affect miRNA activity. Like R495X, P525L causes a severe form of ALS with massive cytoplasmic FUS accumulation. He hopes the mechanistic work reported here can be replicated in motor neurons derived from patient-induced pluripotent stem cells and postmortem samples. A mouse model of the R495X FUS mutation exists, as well, see Research Models.—Marina Chicurel
References
Research Models Citations
Paper Citations
- Deng H, Gao K, Jankovic J. The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol. 2014 Jun;10(6):337-48. Epub 2014 May 20 PubMed.
- Dini Modigliani S, Morlando M, Errichelli L, Sabatelli M, Bozzoni I. An ALS-associated mutation in the FUS 3'-UTR disrupts a microRNA-FUS regulatory circuitry. Nat Commun. 2014 Jul 9;5:4335. PubMed.
Further Reading
Papers
- Rinchetti P, Rizzuti M, Faravelli I, Corti S. MicroRNA Metabolism and Dysregulation in Amyotrophic Lateral Sclerosis. Mol Neurobiol. 2017 Apr 18; PubMed.
Primary Papers
- Zhang T, Wu YC, Mullane P, Ji YJ, Liu H, He L, Arora A, Hwang HY, Alessi AF, Niaki AG, Periz G, Guo L, Wang H, Elkayam E, Joshua-Tor L, Myong S, Kim JK, Shorter J, Ong SE, Leung AK, Wang J. FUS Regulates Activity of MicroRNA-Mediated Gene Silencing. Mol Cell. 2018 Mar 1;69(5):787-801.e8. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.