Is This Mutation Pathogenic? Alzforum Updates System for Making This Call
Quick Links
The Alzforum Mutations database has upped its game. This repository includes rap sheets on more than 500 variants in the three autosomal-dominant Alzheimer’s disease genes amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN 2). To estimate the pathogenicity of each one, the scientists curating this database have now applied a new classification system. Incorporating guidelines from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP), Alzforum curators weigh genetic, clinical, molecular, and functional evidence on each variant, and use an algorithm to predict its pathogenicity. Alzforum encourages our world-wide audience to peruse the new rankings and the methods used to call them, which are extensively explained.
- Alzforum mutation database updates its pathogenicity classification system.
- More than 500 variants in familial AD genes have been reclassified.
- [Editor's note: With this story, Alzforum reporters are reporting on the work of Alzforum curators.]
Over the past 30 years, hundreds of variants in the three ADAD genes have been reported, and for most of them the initial paper claimed they were pathogenic. Alas, since then geneticists have learned that they don’t all pack an equal punch. Some trigger disease at a devastatingly young age, while others may have no effect on their carriers whatsoever, and effects in between these two extremes occur, as well. As mutations have surfaced over the years, Alzforum has cataloged them, determining their pathogenicity based on available research reports at the time. However, as the number of mutations grew, so too did the amount and the complexity of pathogenicity data. It became clear that Alzforum needed a new classification system.
Several schemes have been developed over the years. For example, one considered a number of pathogenic criteria: evidence of co-segregation with disease; the number of individuals affected; the predicted effect of a variant based on where it resided within the gene; and whether the variant altered Aβ production (Guerreiro et al., 2010). Based on the strength of data at each step, this system classified a given variant as definitively, probably, possibly, or not pathogenic, or as a potential risk factor. Though this and other classification systems were a step forward for the field, they were challenging to implement consistently and objectively across more than 500 mutations, and sometimes did not include the most recently available genetic information.
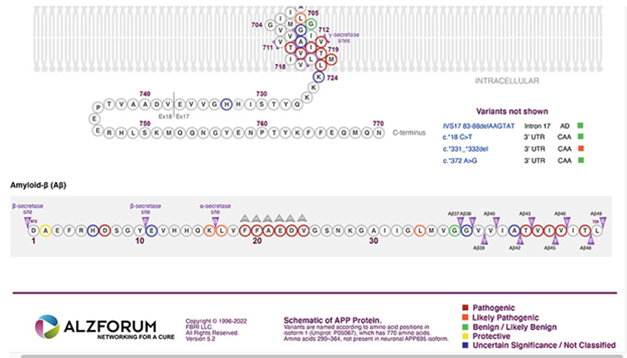
Menagerie of Mutants. In this interactive schematic of variants in APP, readers can click on each variant to explore its pathogenicity, which is classified based on the new, ACMG-based criteria.
Alzforum set out to develop a classification system that could be endorsed by both geneticists and the broader AD research community, using the ACMG-AMP guidelines as a backbone. The ACMG-AMP scheme employs two sets of classification criteria: one assesses pathogenicity, the other benignity. It identifies one standardized criterion, dubbed PVS1, that would constitute very strong evidence of pathogenicity, four criteria that constitute strong evidence (PS1-PS4), six that make for moderate evidence (PM1-PM5), and five as supporting (PP1-PP5). Similarly, one criterion qualifies a variant as stand-alone benign (BA1), the strongest evidence for an innocuous variant, four as strong evidence (BS1-4), and seven as supporting (BP1-7). Furthermore, Alzforum curators upgrade, downgrade, or maintain the weighting of criteria based on available data. This is reflected in a second letter following the ACMG criteria.
How does this work in practice? Take the E693G Arctic variant in APP, for example. Alzforum curators agree that the variant meets the PS3 criteria, which require “well-established in vitro or in vivo functional studies supportive of a damaging effect on the gene or gene product.” Hence E693 gets a PS3-S rating in the Alzforum database, S standing for strong (see image below). The curators bumped up the criteria from supporting to strong in the case of PP1, because of strong co-segregation of the variant with disease, rendering a PP1-S rating. Finally, they maintained the moderate criteria of PM1-M, PM2-M, and PM-5M. Similar calls have been made for hundreds of other mutations in the database.

To sum all these weighted criteria, our curator feeds the rankings into InterVar, an algorithm designed to estimate pathogenicity based on ACMG criteria. The algorithm then renders a final call, classifying the variant as pathogenic, likely pathogenic, benign, or likely benign. If too few criteria are met, or if the benign and pathogenic criteria contradict each other, the pathogenicity is classified as “uncertain significance.”
There are two exceptions to these classifications. Alzforum added a protective category. So far, it includes only the A673T in APP, aka the “Icelandic mutation,” which attenuates Aβ production (Jul 2012 news). And for situations in which data are flimsy or sparse—for example, a variant detected in a single person—Alzforum stopped short of running them through InterVar, opting instead to label such variants as “Not Classified.” More than 200 unclassified variants have been reported in APP, PSEN1, and PSEN2. Despite their status, readers can still peruse all available information about them, including any pathogenicity criteria they may have met thus far.
Also of note, the ACMG-AMP guidelines are designed to classify variants inherited in a Mendelian fashion. The classification of non-Mendelian variants, such as risk factors, is outside their scope. For the few APP, PSEN1, and PSEN2 variants suspected to fall into this latter category, Alzforum notes the limitation under the variant's assigned classification term.
As of January 2022, the new classification system reduced the number of variants Alzforum had previously classified as “pathogenic” from about 72 percent to about 28 percent. Fourteen percent of variants are currently classified as “likely pathogenic,” while 42 percent are “not classified.”
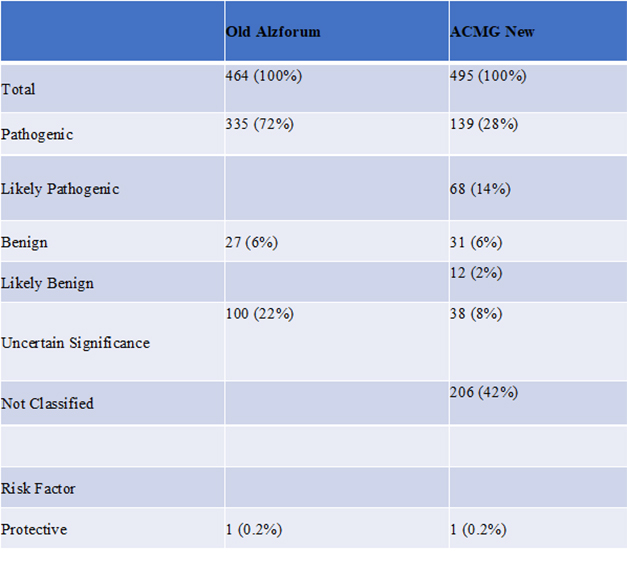
New Classifications. Alzforum’s new ACMG-based pathogenicity analysis added three new categories, cutting the number of variants deemed pathogenic (Jan 2022).
Geneticists surveyed so far agree that the new methodology for classifying variants is better than prior schemes, though some took issue with specific classifications and offered suggestions for changes.
Gaël Nicolas of Normandie University in Rouen, France, called the new system more objective than previous classification schemes. He frequently relies on information in the Alzforum Mutations database to conduct his own classification of variants, and has used the ACMG guidelines for years. That said, he noted that for some variants, determining pathogenicity requires specific knowledge about how those proteins work in the context of AD, and may not fit neatly into the ACMG categorization scheme. He suggested gene-specific types of evidence—such as a mutation that lands within a region encoding the catalytic site of PSEN1—be taken into account. Alzforum curators did incorporate several AD-specific factors into their assessments, though not all of those suggested by Nicolas. He also agreed with some, but not all, classifications of variants in the new system.
“These changes will be extremely useful for us (and for the field), especially when assessing new variants for pathogenicity,” commented Ellen Ziegemeier of Washington University in St. Louis. She is part of the Dominantly Inherited Alzheimer Network, which documents, classifies, and studies hundreds of mutations among its participating families. “We have been updating our guidelines as well, and see that we are in alignment on most of the points,” Ziegemeier wrote.
Are you a clinical geneticist? A human genetics researcher? A genetic counselor who works with families afflicted with multigenerational Alzheimer's? Perhaps you are a molecular biologist working on mutant APP or PS variants? Get acquainted with our new system and offer your input in the comment box below.—Jessica Shugart
References
Book page Citations
Mutations Citations
Mutation Interactive Images Citations
News Citations
Paper Citations
- Guerreiro RJ, Baquero M, Blesa R, Boada M, Brás JM, Bullido MJ, Calado A, Crook R, Ferreira C, Frank A, Gómez-Isla T, Hernández I, Lleó A, Machado A, Martínez-Lage P, Masdeu J, Molina-Porcel L, Molinuevo JL, Pastor P, Pérez-Tur J, Relvas R, Oliveira CR, Ribeiro MH, Rogaeva E, Sa A, Samaranch L, Sánchez-Valle R, Santana I, Tàrraga L, Valdivieso F, Singleton A, Hardy J, Clarimón J. Genetic screening of Alzheimer's disease genes in Iberian and African samples yields novel mutations in presenilins and APP. Neurobiol Aging. 2010 May;31(5):725-31. Epub 2008 Jul 30 PubMed.
Other Citations
Further Reading
No Available Further Reading
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.