CRISPR Gene Editing—Poised to Revolutionize Neuroscience?
Quick Links
Bacteria have been doing it for millennia. Researchers, on the other hand, have only recently learned how to wield CRISPR—a gene-editing tool that bestows the power to delete, add, or toy with the expression of genes at virtually any site in the genome. The magic works in every creature tested, from plants to primates. CRISPR’s flashy reputation has prompted some doubts, such as reported off-target effects, but those are gradually being laid to rest by so-called “CRISPR jocks.”
By now, neuroscientists are on the scent. Two recent studies reported that the method effectively silences genes within neurons, both in hippocampal slice cultures and within the brains of living mice. CRISPR promises to dramatically speed up the generation of transgenic animals. It also may allow researchers to repair mutations in neural stem cells, and possibly facilitate gene therapy in humans. Researchers have already corrected disease-causing mutations in mouse models of muscular dystrophy and a fatal liver disease. CRISPR studies of neurodegenerative diseases are just starting to ramp up, as researchers are using the new technique to build animal models and develop potential therapies.
“CRISPR is a game-changer, and it’s utterly simple to use,” Roger Nicoll of the University of California, San Francisco, told Alzforum. An electrophysiologist who claims to know next to nothing about manipulating genes, Nicoll used CRISPR to delete glutamate receptors in hippocampal neurons, as reported in the September 3 Neuron. “It was the most satisfying, uncomplicated study that I’ve done,” he said.
CRISPR Kaleidoscope. Cas9 nuclease unfurls itself to bring a guide RNA and its target DNA sequence together. Once bound, the enzyme makes a double-stranded DNA break, initiating CRISPR-mediated genome editing.
The acronym CRISPR, for clustered regularly interspaced short palindromic repeats, was first coined in 2002 to describe a phenomenon uncovered by decades of research. Nearly half of all bacterial species and almost all archaeal species whose genomes have been sequenced harbor a curious breed of clustered repeat sequences. These repeats were interspersed between nonrepetitive sequences, which later proved to be snippets of genetic material from phages, the viruses that infect bacteria (see Jansen et al., 2002). Eventually, researchers figured out that bacteria had used these nonrepetitive sequences as a defense against phage infection. When phages invade, the bacteria transcribe these captured DNA sequences, at which point a bacterial nuclease called Cas9 dices up the transcripts and forms complexes with the pieces. Then the RNA fragments pair up with complementary sequences in the invading phage DNA, whereupon Cas9 exacts a double strand break. As these genomic fractures grow in numbers, they ultimately neutralize the phage.
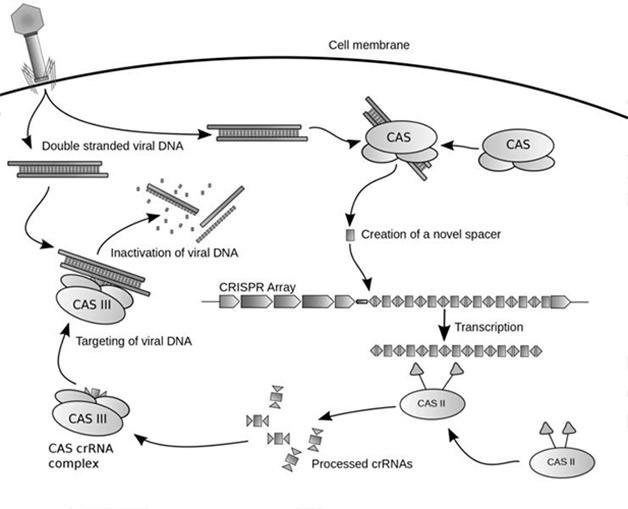
Phage Hitman. Researchers first discovered CRISPR as a bacterial defense system against phage viruses. The Cas9 nuclease snips phage DNA and pockets it in the bacterial genome as novel spacer sequences. The nuclease later transcribes those to hunt down complementary sequences in the phage and dice up its DNA. [Courtesy of James Atmos, Wikimedia Commons.]
Researchers next learned that they could co-opt this system to silence any gene. After double strand breaks occur, the host naturally repairs them using a process called non-homologous end joining. NHEJ is a haphazard type of repair that adds or removes nucleotides to the target sequence to ligate the broken ends, usually causing a frameshift that renders the gene defunct.
Just last year, scientists got the system to work in mammalian cells by transfecting them with appropriate guide RNAs along with the Cas9 gene derived from Streptococcus pyogenes (see Cong et al., 2013, and Mali et al., 2013). Since then, applications have evolved at a blistering pace. Researchers can now use CRISPR to edit or add genes. After transfecting cells with desired “filler” sequences flanked with DNA homologous to the insertion site, cells are coaxed to use homologous recombination, rather than NHEJ, to insert the foreign fillers into the host DNA. Using this more controlled technique, researchers can correct mutations or add new ones, insert reporter sequences, or knock in entire genes. Some researchers even fiddle with gene expression by using the CRISPR complex as a delivery vehicle for transcriptional activators and repressors, rather than as a DNA editor (see Gilbert et al., 2013, and Maeder et al., 2013). The technology eventually will supplant popular gene-editing technologies based on zinc finger and TALEN endonucleases, which are more time-consuming and costly to run than CRISPR, scientists believe. “Right now, it seems that there are no limits to what you can use [CRISPR] for,” Nicoll said.
Nicoll jumped aboard the CRISPR bandwagon to knock out glutamate receptors in neurons within hippocampal slice cultures. His lab had previously used the Cre/Lox system to do this. “The limitation there is, you have to make the whole mouse,” he said. This process can take years if more than one mutation or transgene is desired. “Now CRISPR comes along, and you can make multiple mutations simultaneously in about a month or two, the time it takes you to make the constructs,” Nicoll said.
Co-first authors Salvatore Incontro and Cedric Asensio cut their teeth on CRISPR by targeting the subunits GluN1 and GluA2 of the NMDAR and AMPAR glutamate receptors that are expressed in excitatory synapses. (These are among the receptors that have been proposed to bind Aβ oligomers and mediate neurotoxic consequences in Alzheimer’s disease.) The researchers coated plasmids containing complementary guide GluN1 and GluA2 RNAs, along with a plasmid containing Cas9, onto gold particles, and blasted them onto hippocampal slice cultures using a gene gun. Every transfected neuron they looked at failed to produce excitatory signals when given the proper stimulus, suggesting that their glutamate receptors were gone. The phenotypes were identical to those seen in Cre/Lox-based subunit knockouts.
Another paper, published in PLOS One a day before Nicoll’s, also reported knocking down the same GluN1 subunit in neuronal slice cultures. Led by Bernardo Sabatini at Harvard University, this study used CRISPR in utero. First author Christoph Straub and colleagues injected the CRISPR constructs directly into the brains of mouse embryos 15 days after conception—a time when neuronal progenitors are starting to differentiate into bona fide neurons, Sabatini told Alzforum. Two to three weeks after the pups were born, the researchers measured recordings of hippocampal neurons, and found that every transfected cell was devoid of NMDAR-mediated signals. The in utero CRISPR technique allowed the researchers to knock down a gene in cells within their native developmental context, Sabatini said. He added that the technique offered a refreshing departure from issues known to plague RNA interference experiments, such as incomplete knock down. “We, as a community, are tremendously excited about using CRISPR in neurons, mostly because we have had so much trouble with RNAi,” Sabatini said. “I have no doubt that any lab that has ever done RNAi is trying this.”
To learn how scientists use CRISPR to make transgenic animals, and about early forays into neurodegenerative disease research, read part 2 of this story.—Jessica Shugart
References
News Citations
Paper Citations
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002 Mar;43(6):1565-75. PubMed.
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013 Feb 15;339(6121):819-23. Epub 2013 Jan 3 PubMed.
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013 Feb 15;339(6121):823-6. Epub 2013 Jan 3 PubMed.
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013 Jul 18;154(2):442-51. Epub 2013 Jul 11 PubMed.
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013 Oct;10(10):977-9. Epub 2013 Jul 25 PubMed.
Further Reading
No Available Further Reading
Primary Papers
- Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, Complete Deletion of Synaptic Proteins using CRISPR. Neuron. 2014 Sep 3;83(5):1051-7. Epub 2014 Aug 21 PubMed.
- Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-Mediated Gene Knock-Down in Post-Mitotic Neurons. PLoS One. 2014;9(8):e105584. Epub 2014 Aug 20 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.