Cellular Biosensor Detects Tau Seeds Long Before They Sprout Pathology
Quick Links
Scientists believe that the protein suspects in neurodegenerative diseases use prion-like templating to spread throughout the brain. However, catching these bad seeds in the act has been an enduring challenge. Now, researchers at Washington University in St. Louis led by Marc Diamond have developed a cell-based assay that they claim does the job, and they are poised to share it with other researchers by donating the cells to the ATCC cell repository. The cells fluoresce when mixed with minute amounts of either tau or α-synuclein seeds. Described September 26 in Proceedings of the National Academy of Sciences, the new method detected seeds in mouse models of tauopathy weeks to months before tau pathology was apparent. It revealed seeds in postmortem extracts from AD brains.
The researchers hope the method will facilitate rapid tracking of seeding throughout the animal brain, become a routine measure in human postmortem brain samples, and facilitate the detection of seeds in fluids such as CSF. Diamond, who has just moved to the University of Texas Southwestern Medical Center in Dallas, posted the procedure to the Alzforum protocol database.
“This study adds an important piece of knowledge about the role of tau in neurodegenerative disorders and points to a new direction towards the development of potentially very early preclinical diagnostic tools,” Inga Zerr of Georg-August University in Göttingen, Germany, wrote in an email to Alzforum.
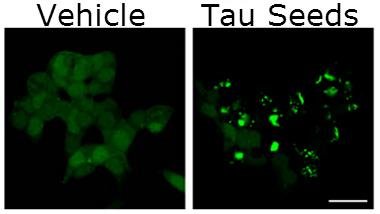
Seed Sensors.
Tau inclusions (green, right) speckle tau biosensor cells one day after treatment with tau fibrils. FRET signals in these cells detect seeds at femtomolar concentrations. [Image courtesy of Holmes et al., PNAS 2014.]
Studies from Diamond’s and other labs have reported that tau pathology, in the form of neurofibrillary tangles and aggregates, can move throughout the brain (see Mar 2009 news story). Toxic tau species are thought to pass from affected to anatomically connected unaffected cells, where the seeds are thought to serve as templates that corrupt normal tau proteins. However, the relationship between these mobile tau species and the progression of disease is unclear (see Aug 2013 conference story).
To better understand that relationship, Diamond’s group had previously developed an assay to detect tau seeds based on fluorescence resonance energy transfer (FRET) (see Kfoury et al., 2012). The method discerns the proximity of two molecules by measuring the transfer of energy from a donor fluorophore attached to one molecule to an acceptor fluorophore on the other. Diamond expressed donor- and acceptor-tagged tau in HEK293 cells, dosed the cells with tau fibrillar seeds, and monitored the subsequent fibrillization, or seeding, of cellular tau via FRET. Initially, the researchers used a micro-well plate reader to quantify the signal and looked at individual cells using fluorescence microscopy to home in on qualitative differences. Both had their disadvantages—the plate reader looked at too many cells at once, masking subtle signals, and microscopy was laborious and could only assess a few cells at time. The researchers stepped up their game. They generated a stable monoclonal cell line expressing FRET-ready constructs that can be analyzed via flow cytometry. This technique allows for both single-cell and population-based analysis, which, when combined, make for a far more sensitive measure of FRET.
Co-first authors Brandon Holmes and Jennifer Furman cloned HEK293 cells that stably expressed the tau repeat domain containing the disease-associated P301S mutation. They tagged the mutant tau with cyan (donor) or yellow (acceptor) fluorescent protein (CFP/YFP). The researchers treated the cells with increasing concentrations of tau fibrils prepared in vitro. Measuring FRET signals in individual cells, they detected seeding activity using as little as 300 femtomolar fibrils. This is about 30,000-fold more sensitive than the assay the group had previously used, Diamond said. The FRET signals increased linearly over three orders of magnitude as more tau seeds were added. This suggests the assay would be useful for comparing samples with vastly different levels of tau seeds, as well as distinguishing between samples with subtle differences.
To confirm that the assay detects bona fide tau seeds, the researchers separated FRET-positive and -negative cells using a fluorescent-cell sorter, and examined them with a confocal microscope. Nearly all the FRET-positive cells contained intracellular tau inclusions indicative of seeding, whereas virtually none of the FRET-negative cells did.
The assay also worked for α-synuclein seeds. Cell lines that expressed CFP/YFP-tagged α-synuclein harboring the disease-associated A53T mutation began fluorescing within one day of being exposed to α-synuclein fibrils made in vitro. Importantly, the biosensor cell lines only emitted FRET signals when stimulated by seeds of their own ilk. Alpha-synuclein seeds did not trigger fibrillization in cells expressing tau reporters, or vice versa.
The researchers next wanted to see how the seeding activity detected by the biosensor cells correlated with traditional tests of tau aggregation. Researchers commonly turn to a suite of monoclonal antibodies that recognize aggregated and/or hyperphosphorylated forms of tau to measure tauopathy. To correlate these with their biosensor, the researchers used transgenic mice expressing human tau with the P301S mutation. These mice develop tau pathology and cognitive impairment by six months. Using the MC1 antibody, which recognizes pathological conformations of tau, the researchers detected the protein in mouse brain sections as early as three months. The AT8 and PG5 antibodies, specific for different hyperphosphorylated forms of tau, detected their targets no earlier than six months, and thioflavin S bound tau fibrils by nine months. However, the biosensor cells picked up tau seeds in 1½-month old mouse brains, consistent with the idea that seeds precede the development of tauopathy. Seeding activity in extracts from the brainstem, neocortex, frontal lobe, and hippocampus of these mice increased by three orders of magnitude by 12 months.
The researchers next tested their biosensor cell lines with postmortem brain samples. Only extracts from AD brains—not from controls or brains of people with Huntington’s disease—triggered a FRET signal. When the researchers depleted the extracts with a tau-specific antibody before dosing the cells, the FRET signal dropped significantly, suggesting that the seeding activity in the extracts derived primarily from tau protein.
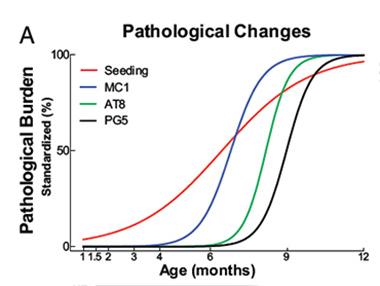
Ahead of the Curve.
Tau biosensor cells detected tau seeds (red line) in P301S transgenic mice several months before traditional histological methods detected pathological forms of tau. [Image courtesy of Holmes et al., PNAS 2014.]
Dominic Walsh of Brigham and Women’s Hospital in Boston commented that the study presented solid findings, but said he would have liked to see more experiments testing the cell lines on human tissues and fluids, rather than transgenic mice. “Ideally with an assay like this, you would hope to apply it to a biofluid such as CSF or blood,” he said.
Holmes and Furman were unable to detect seeds in human CSF using the assay. This could mean that that the assay is not sensitive enough or that the seeds only exist in the brain parenchyma, Diamond said. Researchers led by Mathias Jucker at the German Center for Neurodegenerative Diseases in Tübingen, Germany, were unable to find Aβ seeds in human CSF despite their existence in the brain (see Sep 2014 news story). Diamond hopes that by improving methods of capturing seeds from biofluids, the assay might ultimately detect seeds if indeed they are there.
Walsh noted that researchers do not yet know if the seeds they detect are pathological. “That’s the elephant in the room,” Walsh said. He added, however, that as long as the seeding correlates with disease progression, it could be useful as a diagnostic. “Perhaps the seeding itself parallels a change in some other form of tau that causes disease,” he said.
Bradley Hyman of Massachusetts General Hospital in Charlestown commented that the full role of tau seeding in disease has yet to be appreciated, but that the assay would be useful for doing so. “The work presented here shows nicely that their approach detects changes earlier than previous technologies, and now gives us tools to begin to explore the consequences of those (even earlier) changes,” he said.
Elizabeth Fisher of University College London commented that the findings add to the growing concept of a shared prion-like mechanism for neurodegenerative disease. “We can see that evidence for common prion-like mechanisms in neurodegenerative disease is robust and clear—which has exciting implications for potentially developing related therapeutic strategies for these diseases,” she wrote.—Jessica Shugart
References
Protocol Citations
News Citations
- Double Paper Alert—Keystone Presentations Now in Press
- Tales of Traveling Tau: Is Transfer Between Neurons Normal?
- Bad Seeds—Potent Aβ Peptides Instigate Plaques, Won’t Be Fixed
Research Models Citations
Paper Citations
- Kfoury N, Holmes BB, Jiang H, Holtzman DM, Diamond MI. Trans-cellular propagation of Tau aggregation by fibrillar species. J Biol Chem. 2012 Jun 1;287(23):19440-51. Epub 2012 Mar 29 PubMed.
Further Reading
Papers
- Holmes BB, Diamond MI. Prion-like properties of Tau protein: the importance of extracellular Tau as a therapeutic target. J Biol Chem. 2014 Jul 18;289(29):19855-61. Epub 2014 May 23 PubMed.
- Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010 Mar;11(3):155-9. PubMed.
- Holmes BB, Diamond MI. Cellular mechanisms of protein aggregate propagation. Curr Opin Neurol. 2012 Dec;25(6):721-6. PubMed.
Primary Papers
- Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, Belaygorod L, Cairns NJ, Holtzman DM, Diamond MI. Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):E4376-85. Epub 2014 Sep 26 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Institute of Neurology
Marc Diamond and colleagues at Wash U have produced a very exciting paper underlining the similar “prion-like” mechanisms of common neurodegenerative diseases. The authors have developed an excellent, sensitive assay system for looking at tau seeding in human and mouse tissues. They find clear seeding activity long before any detectable histopathological changes and this is exactly what was shown early this year by John Collinge’s group for the prion protein: Neuropathology only occurs sometime after reaching a particular titre of the misfolded protein (Sandberg et al., 2014). As it happens, David Borchelt’s group has just published the first animal-to-animal transmission of SOD1 pathology, which causes another neurodegenerative disease, amyotrophic lateral sclerosis, and so we can see that evidence for common prion-like mechanisms in neurodegenerative disease is robust and clear (Ayers et al., 2014). This has exciting implications for potentially developing related therapeutic strategies for these diseases.
View all comments by Elizabeth M. FisherThis work, using a FRET-based assay that amplifies the signal from conformationally misfolded proteins, continues a strong series of studies from the Diamond lab examining the cell biology of misfolded tau and its ability to cross cell membranes. Especially interesting here is the development of an assay—and its general applicability—that demonstrates ultrasensitive detection of such misfolded proteins. While a full appreciation will come over time for how this information fits into more traditional approaches (dating back to silver stains that—conceptually at least—detect and visualize abnormally folded proteins), the work presented here shows nicely that the new approach detects changes sooner than previous technologies, and now gives us tools to begin to explore the consequences of those early changes. This is a “must read” paper.
View all comments by Bradley Hyman�
This is an absolutely important paper on the development of an assay that detects tau seeding activity using a combination of FRET flow cytometry and a tau monoclonal FRET biosensor line. By combining the techniques the authors demonstrated they can detect seeds in the femtomolar range. Using brain homogenates from AD patients and people with other neurodegenerative diseases, they confirmed a high specificity of the seeding reaction, since tau seeding activity was detected in AD brains only. Tau seeding activity increased with age in a mouse model of tauopathy. Moreover, tau seeding activity was the earliest marker of disease pathology in the model. Taken together, these results add an important piece of the puzzle of the role of tau in neurodegenerative disorders and open a new direction toward the development of potentially a very early, preclinical diagnostic tool.
View all comments by Inga ZerrMake a Comment
To make a comment you must login or register.