Compact Mouse Brain Atlas: A New Tool for Studying Neurodegeneration?
Quick Links
As advances in optical microscopy and imaging technologies provide better resolution, scientists have built ever more detailed atlases of the brain. This wealth of information comes at a price: huge data sets that are cumbersome to update and manipulate. In the March 5 Nature Neuroscience, researchers led by Hiroki Ueda at the University of Tokyo describe a compact, portable mouse brain atlas. In its pointillist rendition of the brain, each cell is represented by a simple dot. The researchers illustrate how the atlas, together with the technique used to create it, can be used to measure cell numbers and neuronal activity across the brain.
- Using techniques to clear and expand mouse brains, scientists generate a compact brain atlas.
- Each of its 72 million dots represents a single cell.
- The atlas enables brain-wide assessments of the number, distribution, and activities of labeled cells.
“This is an important step forward in analyzing brain structure and connectivity. It will help perform unbiased analyses of the brain,” said Ali Ertürk, Ludwig Maximilians University of Munich. Michel Goedert, MRC Laboratory of Molecular Biology in Cambridge, England, and Nolwen Rey, Paris-Saclay University, Gif-sur-Yvette, France, thought the tool might help scientists study neurodegenerative disease. “Models of human neurodegenerative diseases often develop filamentous inclusions, but not much is known about the death of cells. We know even less about the effects of seeded, prion-like aggregates on neurodegeneration,” wrote Goedert, adding that the new atlas and the atlas-building methods may help address these questions.
Scientists have commonly assembled three-dimensional atlases of the brain by piecing together images of two-dimensional serial sections (e.g., Allen Brain Atlas; Jun 2013 news; Hawrylycz et al., 2012; Mikula et al., 2007). Although providing high-resolution information, these maps often comprise more than 10 terabytes of data, making it difficult to create and examine multiple maps from different individuals. For example, using these atlases to align and compare whole-brain, single-cell data from several animals treated for different times with a drug would be challenging. “These atlases are not scaleable,” said Ertürk. Also, collections of tissue sections are often suboptimal for studying far-flung connections between neurons, he said.
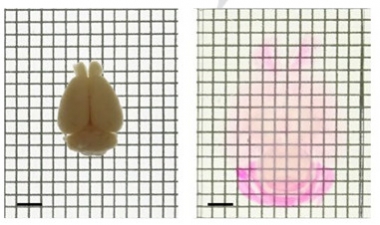
Cleared and Expanded.
A fixed mouse brain (left) becomes enlarged and transparent (right) after treatment with a chemical cocktail. Propidium iodide (pink) then identifies cell nuclei. [Courtesy of Murakami et al., Nature Neuroscience, 2018.]
Hoping to create an atlas to explore the whole brain at once under different conditions, first author Tatsuya Murakami adopted a new imaging technique developed in Ueda’s lab. Clear, Unobstructed Brain Imaging cocktails and Computational analysis, or CUBIC for short, immerses samples in a mixture of amino alcohols that strip away lipids, rendering the tissue transparent (Susaki et al., 2014). Compared with CLARITY, SeeDB, ScaleS, and other tissue-clearing methods, CUBIC avoids long treatment periods, loss of fluorescent signals, and the need for a specialized device to extract lipids (Apr 2013 news; Sep 2015 news; Ke et al., 2013). CUBIC also includes an image analysis tool that aligns images from different brains to compare them with each other. Murakami first cleared whole brains and then stained nuclei using propidium iodide, which binds DNA.
He also searched for compounds that would swell the tissue to more clearly discern individual cells where they are tightly packed, such as in the granular layer of the cerebellum. “We screened more than 1,600 chemicals to identify the best ones to enlarge the brain,” Ueda said. In the modified method, called CUBIC-X, imidazole followed by antipyrine increased the brain volume about 10-fold (image above). “There was no cell loss or apparent distortion,” said Ueda.
The researchers then used a customized, light-sheet fluorescence microscope to visualize the propidium iodide-labeled nuclei at high resolution. This type of microscope evokes fluorescence from a thin plane of tissue, just like confocal microscopes do. Collecting stacks of images taken at 5 μm intervals, and rotating the brain to get images from different angles, the researchers compiled 1.3 million images, amounting to 14 terabytes of data. They then applied an imaging algorithm to hone in on the stained nuclei, creating a three-dimensional map representing 72 million cells. This reduced the size of the data set to a mere three gigabytes. “Almost all atlases are based on image parameters, pixels or voxels, for example,” said Ueda. “Ours is based on single cells, the natural unit of life.” Estimates of total cell numbers in mouse brains have been very variable, noted the authors, ranging from less than 60 million to more than 150 million. Although the authors’ estimate may increase with improved resolution of densely populated brain regions, they considered it particularly robust given the number of mice they used, the number of images from each taken from different angles, and the robustness of the counting algorithm.
Next, the researchers aligned the dots with anatomical data sets from the Allen Brain Atlas, and color-coded each based on anatomical location (see image and video below). In the future, the authors plan to overlay other types of information, such as gene expression data. CUBIC-Atlas can be downloaded at http://cubic-atlas.riken.jp.

Compact Brain Atlas. The CUBIC-Atlas maps single cells in two-dimensional “slices” (left) and three-dimensional video (right, and see below). Colors are based on anatomical annotations from the Allen Brain Atlas. [Courtesy of Murakami et al., Nature Neuroscience, 2018.]
3-D CUBIC-Atlas. [ Video courtesy of Dr. Hiroki R. Ueda, Utokyo/RIKEN.]
CUBIC-X and the CUBIC-Atlas can be used to study the number and function of cells across the brain. Transgenic mice expressing fluorescent proteins driven by cell type-specific or activity-dependent promoters open a wide range of possibilities. For example, scientists could measure brain-wide neuronal loss, microglial distribution, or neuronal activity. And because multiple brains can be readily compared with each other, changes in these parameters could be monitored in animals of different ages or under different conditions. Ueda first applied CUBIC-X and CUBIC-Atlas to study development using simply the propidium iodide stain. The researchers processed the brains of different-aged mice, and then mapped the data onto the atlas, which serves as a reference to align data from multiple animals and make comparisons. As expected, they found cells increased in number in the midbrain during the course of early postnatal development. Surprisingly, however, cell numbers dropped in layers 2/3 of the visual and primary somatosensory cortices. Ertürk said this showed how new discoveries can emerge from unbiased examination of the whole brain, and how data from several mouse brains can be uploaded and easily compared with CUBIC-Atlas.
The authors also mapped patterns of neuronal activity using Arc-dVenus mice, which express an unstable yellow fluorescent protein in the forebrain under the control of the Arc promoter (Tatsuki et al., 2016). The gene turns on when neurons depolarize, then dVenus rapidly degrades, serving as a transient marker of neural activity. The researchers mapped fluorescent images from these mice, obtained under different conditions, onto CUBIC-Atlas. Then they used a clustering algorithm to group cells with similar activity patterns. Interestingly, dorsal cells in the dentate gyrus clustered separately from ventral cells, revealing distinct functional regions. “We were surprised,” said Ueda. “We thought the dentate gyrus would behave homogenously.”
Ueda suggested a similar approach could reveal changes in cell number, cell distribution, or cell activity caused by neurodegenerative disease. Ertürk agreed, noting that most people studying neurodegeneration focus on just one or a few brain regions, potentially missing the big picture. Ertürk would like to see CUBIC-X used to trace neural circuitry, as well, especially long-distance connections, which have proven difficult to track. Pavel Osten, Cold Spring Harbor, New York, however, was less enthusiastic, “The methodologies are quite impressive but, as a resource, I don’t think the value is that dramatic,” he said.
How easy will it be for other scientists to use CUBIC-X and the CUBIC-Atlas? Processing the tissue is straightforward, said Ueda. Setting up the customized microscope is likely to be more challenging, predicted Ertürk, even though the paper lays out clear instructions. Researchers may ultimately benefit from software to navigate the atlas and to upload their own data, a project on which Ueda is currently working.—Marina Chicurel
References
News Citations
- An Ultra-High-Resolution, 3D View of the Human Brain
- Brain Anatomy Revealed With CLARITY
- With ScaleS, You Can See Through the Brain to Behold Synapses
Paper Citations
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, Daly BD, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SG, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012 Sep 20;489(7416):391-9. PubMed.
- Mikula S, Trotts I, Stone JM, Jones EG. Internet-enabled high-resolution brain mapping and virtual microscopy. Neuroimage. 2007 Mar;35(1):9-15. Epub 2007 Jan 16 PubMed.
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, Kiyonari H, Shimizu Y, Miyawaki A, Yokota H, Ueda HR. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014 Apr 24;157(3):726-39. Epub 2014 Apr 17 PubMed.
- Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013 Aug;16(8):1154-61. Epub 2013 Jun 23 PubMed.
- Tatsuki F, Sunagawa GA, Shi S, Susaki EA, Yukinaga H, Perrin D, Sumiyama K, Ukai-Tadenuma M, Fujishima H, Ohno R, Tone D, Ode KL, Matsumoto K, Ueda HR. Involvement of Ca(2+)-Dependent Hyperpolarization in Sleep Duration in Mammals. Neuron. 2016 Apr 6;90(1):70-85. Epub 2016 Mar 17 PubMed.
External Citations
Further Reading
Papers
- Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H. The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron. 2017 Nov 1;96(3):542-557. PubMed.
Primary Papers
- Murakami TC, Mano T, Saikawa S, Horiguchi SA, Shigeta D, Baba K, Sekiya H, Shimizu Y, Tanaka KF, Kiyonari H, Iino M, Mochizuki H, Tainaka K, Ueda HR. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat Neurosci. 2018 Mar 5; PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Paris-Saclay University
This new technique allows the three-dimensional imaging of a whole mouse brain with a single-cell resolution without affecting the integrity of the brain analyzed. Brain organization and connections are maintained, and the analysis at such a powerful resolution does not require sectioning and analysis of slices by sampling methods, allowing unbiased quantifications of cellular populations in the entire brain without requiring time-consuming and tedious analysis by unbiased stereology.
The technique developed and the analyses performed here are truly impressive, culminating in a three-dimensional atlas of the mouse brain with a level of detail and precision that could not be achieved with classical techniques, nor with previous clearing methods.
The technique is proven to be efficient when using fluorescence reporter proteins, but ultimately being able to apply it as well to brains stained with classical immunofluorescence techniques will broaden its applications.
In addition, the current whole-brain analysis of the cell population is based on a nuclear marker that is easier to identify than a marker of the cell body. Hopefully, this technique can be expanded to perform single-cell detection of cellular makers localized within the cytoplasm (more challenging due to the diversity of cell sizes and shapes) that would allow cell quantification of specific cellular populations, or even to track single neuron projections throughout the brain. These techniques would allow faster and easier analysis of cell loss in specific cell populations in the whole brain in neurodegenerative models (but not only) and might help us identify more precisely the extent of axonal degeneration, pathological conditions, and the specific pathways involved in disease models.
MRC Laboratory of Molecular Biology
This paper used whole organ clearing and homogeneous tissue expansion for producing a three-dimensional atlas of the mammalian brain. Nerve cell numbers could be quantitated over time in a given brain region in an unbiased manner. Thus, the authors showed a reduction in nerve cell numbers in the somatosensory cortex and the visual area during early postnatal development. Models of human neurodegenerative diseases often develop filamentous inclusions, but not much is known about the death of cells. We know even less about the effects of seeded, prion-like aggregates on neurodegeneration. Work such as that described here may lend itself to address these important questions.
Make a Comment
To make a comment you must login or register.