GnomAD: Stunning Variation Seen in Largest Collection of Human Genomes
Quick Links
In seven papers in Nature journals published May 27, the Genome Aggregation Database (gnomAD) consortium unleashed analyses of 125,748 exomes and 15,708 whole genomes, hailing from unrelated people living on six continents across the globe. The sheer size of the dataset allowed the most comprehensive analysis of human genetic variation to date.
- Aggregate analyses of 141,456 human genomic sequences published.
- Catalog ranks predicted loss-of-function variants by consequence.
- LRRK2 loss-of-function variants well-tolerated, supporting therapeutic inhibition strategy.
In their summary paper, researchers led by Daniel MacArthur, based at the Broad Institute of MIT and Harvard at the time the work was done and now at the Garvan Institute of Medical Research and Murdoch Children's Research Institute in Australia, unearthed hundreds of thousands of genetic variants predicted to wipe out expression of the proteins they encode, and used the prevalence of the variants to gauge how essential each gene is for human life.
Other scientists leveraged the massive dataset to zero in on variation in a single gene—LRRK2—reporting that people who lack one functional copy of it fare just fine. Ergo, the scientists reason, therapeutic LRRK2 inhibition might not cause untoward consequences. Myriad other trends popped out of the data, including a disturbing diversity of structural variants involving more than 50 nucleotides each, which collectively account for more than a quarter of expression-nixing variants in people’s genomes.
“With this type of analyses, we begin to enter in a phase of deeper understanding of the impact of genetic variations thanks to a long-term quality control of annotations and the largest compendium of DNA sequences,” commented Philippe Amouyel of Institut Pasteur, Lille, France. “These articles offer new hope in this hunt for pathophysiological knowledge, especially in the field of neurodegenerative disease.”
“These papers highlight the value of large-scale human sequencing projects, and in particular, the study of rare predicted loss-of-function variants as a way to assess the likelihood of toxicity being associated with inhibition of a protein by potential novel therapeutics,” wrote Alison Goate of the Icahn School of Medicine at Mount Sinai in New York.
GnomAD is the heftier successor to ExAC. This catalog of more than 60,000 exomes transformed the study of human genetic variation (Aug 2016 news). GnomAD doubles the number of exomes and, importantly, adds whole genome sequences to the compilation. The sequences are the spoils of more than 60 case-control studies of adult-onset disorders, including diabetes, cardiovascular disease, and psychiatric disorders, which shared raw sequencing data with the consortium.
Similar to ExAC, which was available to geneticists several years before its formal publication in 2016, researchers have had access to the raw data from GnomAD’s 125,748 exomes and 15,708 whole genomes for three years already, MacArthur told Alzforum. In that time, members of the consortium have doggedly worked to harmonize and improve the quality of the mind-boggling 3-petabyte dataset (that’s 3 million gigabytes). They have also run large-scale analyses to uncover overarching patterns of functional significance in the human genome. Co-directed by Heidi Rehm and Mark Daly at the Broad Institute at Massachusetts Institute of Technology in Cambridge, the gnomAD consortium comprises about 150 principle investigators from around the world.
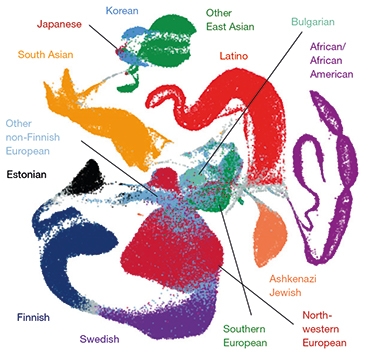
Ancestral Aggregation. A map reflecting the ancestral diversity of the 141,456 people included in gnomAD, using 10 principle components. [Courtesy of Karczewski et al., Nature, 2020.]
In their paper, first author Konrad Karczewski of the Broad Institute at MIT and colleagues described the hunt for predicted loss-of-function variants. These variants snuff out gene expression by introducing a premature stop codon or frameshift, or by bungling the splicing of a gene. Why search for these wet blankets? Essentially, pLoF variants illuminate gene function, akin to knockout studies in animal models. Natural selection weeds out carriers of such variants in genes that are essential to life or a person’s ability to reproduce, so researchers can gauge the essentiality of genes based on the frequency of pLoFs detected in the population, relative to the expected number based on the mutation rate of the genome.
Sifting through protein coding sequences in gnomAD, Karczewski discovered 443,769 pLoF variants among 16,694 genes. On average, the researchers detected 18 pLoF variants per gene, and 72 percent of genes had more than 10 pLoF variants. Based on the frequency of variants detected for each gene, the researchers placed genes on a so-called constraint spectrum. It ranged from unconstrained genes, i.e., ones that tolerate pLoF variants, to highly constrained or “intolerant” genes, which were highly depleted from the dataset.
Among genes at the unconstrained end of the spectrum were nonessential genes, or those, like olfactory receptors, that exist in hundreds of versions. Genes at the other end were indispensible in some way. They included those known to be lethal when knocked out of a mouse, or to be necessary to keep human cells alive in a dish, or known haploinsufficient genes, which require two functional copies to support life. These indispensible genes had more contacts within protein interaction networks, and were more likely to be widely expressed across tissues.
Karczewski’s study focused on pLoF variants only within protein coding regions of genes. By their nature, such variants are rare but severe. By contrast, pLoF variants in noncoding regions, as typically emerge in genome-wide association studies, are more common and tend to exert milder effects. Might there be a connection between the two? To find out, the researchers examined GWAS for 658 traits and diseases, and asked whether variants linked to these traits related to the pLoF tolerance of nearby genes. Indeed, the gnomAD researchers found that hits in multiple GWAS commonly mapped near pLoF-intolerant genes. This makes sense, MacArthur said, because even small changes in the expression of these indispensible genes will likely come with consequences, such as increased risk for disease.
OK, you know what gnomAD is. Time now to address the elephant in the room, at least to the mind of the neurodegenerative disease researcher: What about diseases of brain aging? Alas, the spectrum of tolerance to pLoF variation derived from gnomAD is based on how essential a gene is for life and reproductive fitness. Therefore, this type of analysis is largely blind to phenotypes that arise later in life.
Even so, two of the gnomAD companion papers made use of the pLoF studies to try to inform drug-development strategies for diseases of aging. Many drugs aim to inhibit the function of problematic proteins. Similar to the way animal knockouts help scientists estimate the consequences of doing that, pLoF data may come in handy in gauging the feasibility and safety of targeting a gene.
In one paper, first author Eric Minikel started by asking a simple question: Are genes targeted by approved drugs more likely to be tolerant or intolerant to loss of function? To find out, the researchers compared the degree of pLoF tolerance of 383 targets of approved drugs listed in DrugBank with more than 17,000 other protein-encoding genes. They found that genes targeted by drugs were slightly more constrained than non-targeted genes, although they ran the gamut from tolerant to indispensible. The finding contradicts the idea that targeting products of constrained genes is inherently unwise. In fact, Minikel found that 19 percent of these established drug targets were even more intolerant to loss of function than haploinsufficient genes, which are highly constrained. For example, the targets of statins, NSAIDs, and certain chemotherapies are among the most heavily constrained genes.
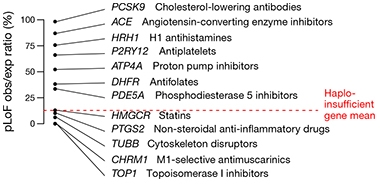
Span the Spectrum. Widely used drugs target the product of genes of different pLoF tolerance. They range from unconstrainted (PCSK9) to highly constrained (TOP1). The mean constraint of haploinsufficient genes (red dashed line) is a proxy for highly constrained genes. [Courtesy of Minikel et al., Nature, 2020.]
Next, Minikel et al. compared the pLoF tolerance of genes encoding proteins that are implicated for their gain-of-function behavior in neurodegenerative diseases, and for which therapeutic inhibitors or suppressors are currently being developed. They are huntingtin (Htt), tau (MAPT), prion protein (Prnp), SOD1, α-synuclein (SNCA), and LRRK2. As with the FDA-approved drugs, these target genes ranged across the entire spectrum of pLoF tolerance.
The most freewheeling gene was Prnp, for which pLoF variants in the N terminus of the gene were completely unconstrained, popping up as often as would be expected by random mutation. Numerous disease-causing, gain-of-function variants clustered in the C-terminus of the protein, though they were collectively almost three times rarer than the pLoF variants in the gene. LRRK2 was slightly constrained, followed by SOD1, HTT, SNCA, and MAPT. For the latter two genes, nary a single pLoF variant was identified in gnomAD, suggesting that having two functional copies of these genes is essential for life.
Oddly, SNCA and MAPT knockout mice are viable and live normal lifespans, although both knockouts have some detrimental phenotypes. APP-Tg mice missing one or both copies of tau avoided memory problems caused by Aβ accumulation (May 2007 news).
HTT was also considered highly constrained, with pLoF variants occurring at 8.2 percent of the frequency expected by chance. This suggests some benefit of carrying two copies of Htt, although previous studies have reported disorders only in cases where two functional copies of the gene were missing (Duyao et al., 1995; Rodan et al., 2016; Ambrose et al., 1994).
Does this mean that therapeutically targeting highly constrained gene products like Htt, α-synuclein, or tau is a dangerous proposition? Not necessarily, suggested the authors. These genes may play essential roles early in development, but could become amenable to targeting later on. Thus far, no early stage clinical trials of α-synuclein or tau inhibition have reported serious side effects. While pLoF tolerance reflects selective pressure on heterozygotes, drugs can be adjusted to inhibit their targets only partially.
Furthermore, Minikel noted that without extensive health data on carriers of pLoF variants, it is difficult to surmise their true impact. That is exactly what first author Nicola Whiffin of Imperial College London and colleagues did for LRRK2 pLoF variants.
Mutations in LRRK2 are strongly tied to Parkinson’s disease, and LRRK2 kinase activity is elevated in people with PD, even among those who do not carry LRRK2 mutations (Di Maio et al., 2018). Several LRRK2 kinase inhibitors and suppressors are in clinical development (DNL201; DNL151; BIIB094). However, worrying phenotypes of LRRK2 knockout mice, or in animals dosed with LRRK2 inhibitors, have caused concern (Hinkle et al., 2012; Fuji et al., 2015; Apr 2020 news).
To investigate how humans handle loss of LRRK2 function, Whiffin and colleagues searched for pLoF variants in the gene in gnomAD, as well as in the 46,062 exome-strong UK Biobank, and in 23andMe, which contains genotype data on more than 4 million customers. Among these three databases, they identified 134 unique pLoF variants among 1,455 carriers, translating into about one in 500 people carrying a LRRK2 LoF variant. For six of the variants, which represent 82.5 percent of the carriers, the researchers confirmed that indeed, LRRK2 expression was significantly reduced.
How did people fare with only one functional copy of LRRK2? The researchers took advantage of different phenotypic data from each database to address this question. For gnomAD and 23andMe, the researchers noted a similar age distribution of LRRK2 pLoF carriers and noncarriers, hinting that loss of LRRK2 function did not dramatically alter lifespan. A subset of the gnomAD carriers had health data available from previous case-control studies in which they had participated, and the researchers found no obvious health problems in carriers compared with noncarriers. Customers of 23andMe fill out extensive health questionnaires, and again, no problems were overrepresented in carriers. The most extensive phenotype data came from UK Biobank, which includes sampling of serum and urine proteins, electronic health records, and death certificates, to name a few. Again, LRRK2 pLoF carriers were no different than noncarriers in any of these rubrics.
Whiffin concluded that lifelong systemic reduction in LRRK2 did not discernably affect health or lifespan, suggesting that LRRK2 inhibitors are unlikely to result in severe issues. The results are consistent with promising safety results of initial trials, and suggest that phenotypes observed in rodent studies may not translate to humans, the authors wrote.
Goate called these findings encouraging, noting that phenotypes previously associated with LRRK2 knockout or inhibition in animal models were not observed in human carriers of pLoF variants in the gene. “However, as the authors point out, carrying a pLoF from conception is not the same as using an inhibitor in later life,” she added. “Despite this caveat, these results provide cautious optimism regarding LRRK2 inhibitors as a treatment for PD.”
Mark Cookson, National Institutes of Health, Bethesda, Maryland, noted that even this dataset is not large enough to test whether losing some LRRK2 protects against Parkinson’s disease, but it does suggest that a partial reduction is at least tolerated throughout life (see full comment below).
Variety of Variants
Other studies in this flurry of new publications used gnomAD to venture beyond the relatively better-charted territory of protein-coding variants into the Wild West of upstream open reading frames (ORFs). First author Whiffin and colleagues reported on this in Nature Communications. Variants in these untranslated stretches that set the gene expression machinery on track can have an outsize impact on gene expression, she found.
Among the 15,708 whole genome sequences in gnomAD, the researchers found 145,398 single-nucleotide variations that either create new start codons, or disrupt stop codons, in uORFs. Essentially, these uORF variants stifle gene expression at the level of translation by creating overlapping open reading frames that snag ribosomes, keeping them from translating the proper downstream gene. These uORF variants were under strong selective pressure, especially if they resided upstream of pLoF-intolerant genes. In essence, this study defined a previously underappreciated category of genetic variants that rival protein-coding pLoFs in their impact.
Taking their own foray into the noncoding abyss of the human genome, researchers led by Michael Talkowski at the Broad charted structural variants, rearrangements of DNA segments involving at least 50 nucleotides. These jumbo variants often elude the gaze of gene sleuths, especially those who use short-read sequencing approaches. First author Ryan Collins and colleagues deployed a powerful mix of computational algorithms to hunt for SVs of six different flavors: deletions, duplications, multiallelic copy number variants, insertions, inversions, and translocations. They also searched for more complex, exotic species, such as those that combine duplications and inversions. They were in for a wild ride.

It’s a Zoo! The varieties of structural variants (and their abbreviations) that researchers uncovered in gnomAD. [Courtesy of Collins et al., Nature, 2020.]
The scientists found 433,371 structural variants lurking in gnomAD, including more than 5,000 of a complex variety. At a whopping 7,439 structural variants per genome—yes, per person on average, so that would be you—the haul more than doubled the number of structural variants identified in previous studies.

Variant Haul. The number of structural variants discovered in gnomAD dwarfs findings from previous studies, listed below. Colors represent variant type (DEL: deletion; DUP: duplication; MCNV: multiallelic copy number variant; INS: insertion; INV: inversion; CPX: complex; BND: breakends). [Courtesy of Collins et al., Nature, 2020.]
More than 90 percent of these variants were rare, occurring at a frequency of less than 1 percent. Half were unique, i.e. were detected only once in the entire dataset.
By analyzing the proximity of structural variants near or within genes, the researchers estimated that structural variants account for more than a quarter of gene inactivation events per genome. They also mess with gene expression more subtly by interfering with regulatory elements that reside in noncoding regions. Nearly 4 percent of genomes analyzed in gnomAD harbored one mega-variant, that is, a DNA rearrangement greater than 1 megabase in size. Finally, the researchers found that 0.32 percent of the genomes had a structural variant predicted to preclude expression of a gene linked to disease. In all, the findings make clear that an astounding menagerie of structural variants exist in people, and hold sway over expression of their genes.
“GnomAD provides a unique opportunity for enhancing our understanding of multiple forms of genomic variation in diverse populations,” commented Jennifer Yokoyama of the University of California, San Francisco. “In addition to illuminating tolerance of loss-of-function variants throughout the genome, the field now has a robust reference for structural variants. This forms the foundation upon which the role of structural variation in neurodegenerative disease can be comprehensively assessed,” she wrote.
In an accompanying editorial in Nature, Deanna Church of Inscripta, Inc., in Boulder, Colorado, hailed gnomAD as an invaluable resource. Church (no relation to George) noted that even more discoveries will emerge with ever-larger datasets. “The consortium’s work has revealed how much information about human variation we had been missing, and has provided tools that help us to better understand the genome at both the population and individual level,” wrote Church. “I can’t wait to see what comes next.”—Jessica Shugart
References
News Citations
- Flood of Exomes Brings Genetic Variation into Focus
- APP Mice: Losing Tau Solves Their Memory Problems
- Sigh of Relief? Lung Effects of LRRK2 Inhibitors are Mild.
Research Models Citations
Therapeutics Citations
Paper Citations
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, Ge P, Vonsattel JP, Gusella JF, Joyner AL. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995 Jul 21;269(5222):407-10. PubMed.
- Rodan LH, Cohen J, Fatemi A, Gillis T, Lucente D, Gusella J, Picker JD. A novel neurodevelopmental disorder associated with compound heterozygous variants in the huntingtin gene. Eur J Hum Genet. 2016 Dec;24(12):1826-1827. Epub 2016 Jun 22 PubMed.
- Ambrose CM, Duyao MP, Barnes G, Bates GP, Lin CS, Srinidhi J, Baxendale S, Hummerich H, Lehrach H, Altherr M, Wasmuth J, Buckler A, Church D, Housman D, Berks M, Micklem G, Durbin R, Dodge A, Read A, Gusella J, MacDonald ME. Structure and expression of the Huntington's disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet. 1994 Jan;20(1):27-38. PubMed.
- Di Maio R, Hoffman EK, Rocha EM, Keeney MT, Sanders LH, De Miranda BR, Zharikov A, Van Laar A, Stepan AF, Lanz TA, Kofler JK, Burton EA, Alessi DR, Hastings TG, Greenamyre JT. LRRK2 activation in idiopathic Parkinson's disease. Sci Transl Med. 2018 Jul 25;10(451) PubMed.
- Hinkle KM, Yue M, Behrouz B, Dächsel JC, Lincoln SJ, Bowles EE, Beevers JE, Dugger B, Winner B, Prots I, Kent CB, Nishioka K, Lin WL, Dickson DW, Janus CJ, Farrer MJ, Melrose HL. LRRK2 knockout mice have an intact dopaminergic system but display alterations in exploratory and motor co-ordination behaviors. Mol Neurodegener. 2012;7:25. PubMed.
- Fuji RN, Flagella M, Baca M, Baptista MA, Brodbeck J, Chan BK, Fiske BK, Honigberg L, Jubb AM, Katavolos P, Lee DW, Lewin-Koh SC, Lin T, Liu X, Liu S, Lyssikatos JP, O'Mahony J, Reichelt M, Roose-Girma M, Sheng Z, Sherer T, Smith A, Solon M, Sweeney ZK, Tarrant J, Urkowitz A, Warming S, Yaylaoglu M, Zhang S, Zhu H, Estrada AA, Watts RJ. Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci Transl Med. 2015 Feb 4;7(273):273ra15. PubMed.
Further Reading
No Available Further Reading
Primary Papers
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O'Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database Consortium, Neale BM, Daly MJ, MacArthur DG. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020 May;581(7809):434-443. Epub 2020 May 27 PubMed.
- Whiffin N, Armean IM, Kleinman A, Marshall JL, Minikel EV, Goodrich JK, Quaife NM, Cole JB, Wang Q, Karczewski KJ, Cummings BB, Francioli L, Laricchia K, Guan A, Alipanahi B, Morrison P, Baptista MA, Merchant KM, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Ware JS, Havulinna AS, Iliadou B, Lee JJ, Nadkarni GN, Whiteman C, 23andMe Research Team, Daly M, Esko T, Hultman C, Loos RJ, Milani L, Palotie A, Pato C, Pato M, Saleheen D, Sullivan PF, Alföldi J, Cannon P, MacArthur DG. The effect of LRRK2 loss-of-function variants in humans. Nat Med. 2020 Jun;26(6):869-877. Epub 2020 May 27 PubMed.
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, Watts NA, Solomonson M, O'Donnell-Luria A, Baumann A, Munshi R, Walker M, Whelan CW, Huang Y, Brookings T, Sharpe T, Stone MR, Valkanas E, Fu J, Tiao G, Laricchia KM, Ruano-Rubio V, Stevens C, Gupta N, Cusick C, Margolin L, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Taylor KD, Lin HJ, Rich SS, Post WS, Chen YI, Rotter JI, Nusbaum C, Philippakis A, Lander E, Gabriel S, Neale BM, Kathiresan S, Daly MJ, Banks E, MacArthur DG, Talkowski ME. A structural variation reference for medical and population genetics. Nature. 2020 May;581(7809):444-451. Epub 2020 May 27 PubMed.
- Minikel EV, Karczewski KJ, Martin HC, Cummings BB, Whiffin N, Rhodes D, Alföldi J, Trembath RC, van Heel DA, Daly MJ, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Schreiber SL, MacArthur DG. Evaluating drug targets through human loss-of-function genetic variation. Nature. 2020 May;581(7809):459-464. Epub 2020 May 27 PubMed. Correction.
- Whiffin N, Karczewski KJ, Zhang X, Chothani S, Smith MJ, Evans DG, Roberts AM, Quaife NM, Schafer S, Rackham O, Alföldi J, O'Donnell-Luria AH, Francioli LC, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Cook SA, Barton PJ, MacArthur DG, Ware JS. Characterising the loss-of-function impact of 5' untranslated region variants in 15,708 individuals. Nat Commun. 2020 May 27;11(1):2523. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Institute Pasteur de Lille
The gnomAD, a new experimental model and a potential useful tool for ND genomics
Several articles, recently published in Nature, Nature Medicine, and Nature Communication, gave an overview of the powerful potential of the Genome Aggregation Database to better understand genetic variations. The Genome Aggregation Database (gnomAD) is a resource developed in the context of an international collaboration whose goal is to aggregate and harmonize exome and whole genome sequencing data from a large number of large-scale sequencing projects, offering this invaluable resource to the scientific community
The V2 release of this database spans 125,748 exome sequences and 15,708 whole-genome sequences from 141,456 unrelated humans sequenced as part of various disease-specific and population genetic studies. This database has been deeply mined by several teams of scientists to begin to express part of its discovery potential : 443,769 high-confidence predicted loss-of-function variants allowing researchers to classify human protein-coding genes along a spectrum representing tolerance to inactivation (Karczewski et al., 2020); a roadmap for “human knockout “ studies that should guide the interpretation of loss-of-function variants in drug development (Minikel et al., 2020); 433,371 structural variations for medical and population genetics (Collins et al., 2020); 1,792,248 multinucleotide variants (Wang et al., 2020); the characterization of the loss-of-function impact of 5' untranslated region variants (Wiffin et al., 2020).
With this type of analyses, we begin to enter into a phase of deeper understanding of the impact of genetic variations thanks to a long-term quality control of annotations and the largest compendium of DNA sequences. For decades we have played with the Cyrillic alphabet, now we begin to read and understand the bible in the Russian language. The main interest of such a large database is to identify rare variations that may have a biological effect, understand not only the impact of these variations on the protein structure itself, but also on its regulatory elements.
Indeed, when we perform Genome Wide Association Studies (GWAS), we identify numerous single-nucleotide polymorphisms that point to a chromosomal region associated with a neurodegenerative disease. However this is only the beginning of the story, because with a position obtained from a statistical test, we do not have any idea of the function of these susceptibility loci in the pathophysiology of the disease. Once we have accumulated all these susceptibility loci by aggregating more and more studies to increase the statistical power to identify rare variants, the experimental evidence needed to characterize the function may take years and years. And among 45 loci you identified, what locus will you experimentally analyze first? We have very powerful high-throughput discovery tools, but we are still lacking high-throughput functional in silico studies to accelerate the understanding of physiopathology, the only way to invent new treatments.
These articles open new hope in this hunt for pathophysiological knowledge, especially in the field of neurodegenerative disease. We can reanalyze our GWAS hits, explore the impact of the mutations on the nearby gene function through a mutational constrained spectrum, and maybe validate new therapeutic drug targets. The identification of new structural variations may help us to decipher the hidden heritability that captivates so many scientists involved in chronic disease genomic research.
The only way to progress in all these domains and to face the always growing complexity of biological systems involved in the pathophysiology of ND, is to continue to develop such huge databases, to facilitate public access to summary data, and to implement global collaborations for the highest benefit of our patients.
References:
Wang Q, Pierce-Hoffman E, Cummings BB, Alföldi J, Francioli LC, Gauthier LD, Hill AJ, O'Donnell-Luria AH, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Karczewski KJ, MacArthur DG. Landscape of multi-nucleotide variants in 125,748 human exomes and 15,708 genomes. Nat Commun. 2020 May 27;11(1):2539. PubMed.
National Institute on Aging
The overall collection of gnomAD papers is an important resource for the field. That the underlying data is in the public domain is really important—it’s a common first stop to evaluate whether disease variants are rare and more likely to be pathogenic, so the resource is important in neurodegenerative disease research.
We also know that some of the specific findings are highly robust. For example, Whiffen et al. examine loss-of-function variants in LRRK2, similar to the lack of difference in frequency between controls and PD cases that we have previously reported (Blauwendraat et al., 2018). Neither study is big enough to say whether this partial loss of LRRK2 is protective against Parkinson’s disease, but both indicate that a 50 percent reduction in expression of this key PD gene is tolerated throughout lifetime.
References:
Blauwendraat C, Reed X, Kia DA, Gan-Or Z, Lesage S, Pihlstrøm L, Guerreiro R, Gibbs JR, Sabir M, Ahmed S, Ding J, Alcalay RN, Hassin-Baer S, Pittman AM, Brooks J, Edsall C, Hernandez DG, Chung SJ, Goldwurm S, Toft M, Schulte C, Bras J, Wood NW, Brice A, Morris HR, Scholz SW, Nalls MA, Singleton AB, Cookson MR, COURAGE-PD (Comprehensive Unbiased Risk Factor Assessment for Genetics and Environment in Parkinson’s Disease) Consortium, the French Parkinson’s Disease Consortium, and the International Parkinson’s Disease Genomics Consortium (IPDGC). Frequency of Loss of Function Variants in LRRK2 in Parkinson Disease. JAMA Neurol. 2018 Nov 1;75(11):1416-1422. PubMed.
King's College London
GnomAD is an essential resource for anyone with an interest in ALS genetics, particularly those researchers like me who are focused on the search for new, highly penetrant dominant pathogenic mutations in our patient cohorts.
The initial release was of limited value to us, as it incorporated variants from the exomes of about 3,000 ALS patients, but this was soon rectified when the “non-neurological” subset of variants from over 115,000 individuals free of neurological conditions became available. Being able to assess accurately the frequency (or novelty) of patient-derived variants in such a large sample of controls, from a diverse group of populations, is a fantastic aid to selecting and prioritizing candidate mutations and genes.
The very recent release of gnomAD version 3, with whole genome variants from over 70,000 individuals, means that we will now be able to assess the frequency of intronic and intergenic variants, regions that have often been neglected in ALS research, with the same accuracy as has been previously been applied to the coding portion of the human genome.
University of Bresica
VIB, University of Antwerp, Center for Molecular Neurology
VIB, University of Antwerp, Center for Molecular Neurology
We also have examined a LRKK2 frameshift variant (c.6187_6191delCTCTA; p.L2063fs*) in lymphoblastoid cell lines (LCLs) of an individual affected by amnestic MCI, without Parkinson’s disease (Perrone et al., 2018). The variant was a five-base-pair deletion in LRRK2 that predicted a frameshift and a premature termination codon after amino acid residue 2063 and was previously reported in one patient with Parkinson’s disease and two control individuals (Ross et al., 2011).
We analyzed the expression at both protein and transcript levels and showed that this LRRK2 mutation had little effect on transcript levels but seemed to result in a nearly complete protein loss in LCLs of the patient carrier.
We further investigated LRRK2 protein levels in control individuals without any LRRK2 mutations and observed a highly variable expression. Some individuals showed near null LRRK2 expression, comparable to the LRRK2 loss observed in the patient carrier. Our protein expression results are slightly different from the ones performed by Whiffin et al., though we also concluded that a low LRRK2 expression is unlikely to interfere with normal biological processes.
References:
Perrone F, Cacace R, Van Mossevelde S, Van den Bossche T, De Deyn PP, Cras P, Engelborghs S, van der Zee J, Van Broeckhoven C. Genetic screening in early-onset dementia patients with unclear phenotype: relevance for clinical diagnosis. Neurobiol Aging. 2018 Sep;69:292.e7-292.e14. Epub 2018 May 9 PubMed.
Ross OA, Soto-Ortolaza AI, Heckman MG, Aasly JO, Abahuni N, Annesi G, Bacon JA, Bardien S, Bozi M, Brice A, Brighina L, Van Broeckhoven C, Carr J, Chartier-Harlin MC, Dardiotis E, Dickson DW, Diehl NN, Elbaz A, Ferrarese C, Ferraris A, Fiske B, Gibson JM, Gibson R, Hadjigeorgiou GM, Hattori N, Ioannidis JP, Jasinska-Myga B, Jeon BS, Kim YJ, Klein C, Kruger R, Kyratzi E, Lesage S, Lin CH, Lynch T, Maraganore DM, Mellick GD, Mutez E, Nilsson C, Opala G, Park SS, Puschmann A, Quattrone A, Sharma M, Silburn PA, Sohn YH, Stefanis L, Tadic V, Theuns J, Tomiyama H, Uitti RJ, Valente EM, van de Loo S, Vassilatis DK, Vilariño-Güell C, White LR, Wirdefeldt K, Wszolek ZK, Wu RM, Farrer MJ, Genetic Epidemiology Of Parkinson's Disease (GEO-PD) Consortium. Association of LRRK2 exonic variants with susceptibility to Parkinson's disease: a case-control study. Lancet Neurol. 2011 Oct;10(10):898-908. Epub 2011 Aug 30 PubMed.
Make a Comment
To make a comment you must login or register.