Method of the Year—Microscopy Advances Take Biology by STORM
Quick Links
Super high-resolution light microscopy won, hands-down, Nature magazine’s 2008 method of the year designation. This set of techniques encompasses techniques such as STORM (stochastic optical reconstruction microscopy), STED (stimulated emission depletion), and PALM (photoactivation localization microscopy), and together they have made possible the resolution of spots much as described in a special feature in the December 17 Nature Methods online. These new “nanoscopies” allow researchers to see structural, and even molecular, details previously impossible to resolvable in living cells. They also allow multiplex labeling with high molecular specificity. “This exciting prospect has driven our choice of nanoscopy as Method of the Year 2008,” write Nature’s editors. Alzforum has covered some of these advances as they were published, particularly those that are relevant to neurobiology. We reported how STED was used to support the “kiss and run” theory of synaptic vesicle recycling (see ARF related news story) and how these advanced techniques can be used to reveal intricate 3D structures, such as the nuclear pore complex (see ARF related news story). A video explaining the Method of the Year is freely viewable at Nature.com.
The bane of light microscopy has always been diffraction, which blurs images and effectively limits resolution to about half the wavelength of the radiation being used. For visible light that means about 200 - 250 nm. This diffraction barrier was predicted by such luminaries as Lord Rayleigh and Émile Verdet in the latter half of the nineteenth century, and mathematically described by Ernst Abbe in 1873. Has it finally been broken? Not quite. What all these newer types of microscopy do is to separate very close objects (less than 200 nm apart) not in space, but in time, sequentially turning on and off different fluorophores. It is not so much that nanoscopies break the diffraction barrier as that they cleverly tap-dance around it.
In fact, as nanoscopy pioneer Stefan Hell from the Max Planck Institute for Biophysical Chemistry in Goettingen, Germany, writes in a Nature perspective, “Discerning objects or molecules with distinct spectral characteristics has never been precluded by diffraction.” In other words, when light of two distinct colors is used to illuminate two objects then the diffraction limit does not apply. That’s because filters can simply take care of any diffractive interference. The problem comes when trying to resolve hundreds or thousands of identical, or even different, molecules. There aren’t enough wavelengths to go around. “So if someone had figured out an effective way to label each little object in a sample with a different color, the recent developments would have been less essential,” writes Hell.
Instead, microscopy pioneers have focused on temporal resolution. Though the plethora of emerging nanoscopic techniques differ slightly in approach, they all follow this same principle of sequentially capturing light from different objects. “This is exactly how current far-field optical nanoscopy techniques resolve objects that are closer together than the diffraction limit,” wrote Hell.
Unlike the electron microscope, light nanoscopy is perfectly suited to the study of living cells. Now that resolution is approaching 20 nm—the scale of many macromolecules—the technique will be applicable to various areas of cell biology. In addition to the nuclear pore complex and synaptic vesicle recycling applications mentioned above, nanoscopy has revealed protein clusters in synaptic zones (see Kittel et al., 2006), protein complexes on microtubules (see Huang et al., 2008), lysozomes and mitochondria (Betzig et al., 2006) and even the interaction of mitochondria with microtubules (see image below and Huang et al., 2008).
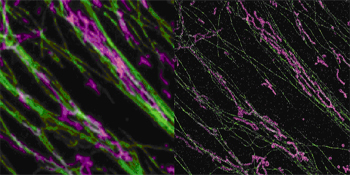
Nanoscopy and Microtubules
A comparison of standard light microscopy (left) versus STORM (right), shows that the latter resolves the interaction between microtubules (green) and mitochondria (pink). Image credit: Xiaowei Zhuang, Harvard University
To be sure, there are likely to be limitations to super-resolution (SR) microscopy. As Jennifer Lippincott-Schwartz writes in a Nature commentary, these include noise from cellular autofluorescence, shortcomings of fluorescent probes, and challenges in labeling specific molecules of interest (antibody-based labeling is inefficient and raises questions about the precise location of the fluorophore, e.g.). She stresses that data from SR microscopy will have to be validated by other means before it becomes widely accepted—a hurdle that electron microscope initially faced, as well. But Lippincott-Schwartz also thinks that scientists can expect to see SR microscopy applied to several predictable areas of biology and also to unpredictable ones. In the former, she includes cellular architecture, heterogeneous molecular organization, and dynamic protein assembly. All three are of interest to researchers studying the molecular pathology that drives Alzheimer and other neurodegenerative diseases. “In the hands of creative biologists, this new extension of the human senses should help uncover many of nature’s secrets,” she concludes.—Tom Fagan.
References
News Citations
- Kiss and Tell—STED Microscopy Resolves Vesicle Recycling Question
- Video of the Week—A New Dimension to Light Microscopy
Paper Citations
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006 May 19;312(5776):1051-4. PubMed.
- Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008 Feb 8;319(5864):810-3. PubMed.
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006 Sep 15;313(5793):1642-5. PubMed.
- Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008 Dec;5(12):1047-52. PubMed.
External Citations
Further Reading
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.