Proteomics Uncovers Potential Markers of Early Autosomal Dominant AD
Quick Links
Recent proteomic studies have offered glimpses into the biology of early Alzheimer’s disease. Two new preprints take this approach further, describing high-throughput surveys that turned up numerous new candidate biomarkers. In one, posted to medRxiv on January 13, scientists led by Carlos Cruchaga at Washington University in St. Louis reported 125 proteins in the CSF that distinguished people who had inherited a familial AD mutation from relatives who had not. Together, nine of these proteins differentiated carriers from noncarriers better than did established CSF Aβ and tau biomarkers. Many of these carriers, from the Dominantly Inherited Alzheimer Network cohort, were years or decades away from developing symptoms.
- In DIAN, largest CSF proteomic study to date finds 125 potential markers.
- A set of nine predicted the presence of FAD mutations better than did CSF Aβ or tau.
- Extracellular matrix protein SMOC1 could be the earliest biomarker.
- In sporadic AD, plasma analysis found completely different proteins.
“Studies such as this one … provide concrete evidence that AD-associated protein dysfunction, beyond Aβ and tau, occurs up to 30 years prior to symptom onset,” Eleanor Drummond at the University of Sydney wrote to Alzforum.
Meanwhile, researchers led by Asim Siddiqui at biotech Seer, Inc., in Redwood City, California, and Steven Arnold at Massachusetts General Hospital, Boston, focused on plasma proteins in people with sporadic AD. In a January 8 bioRxiv preprint, they reported levels of 138 correlated with disease, including eight that associated with cognitive decline. These had little overlap with the DIAN CSF set, indicating that biomarkers may be specific to plasma or CSF, or to disease type.

Candidate Biomarkers. A CSF proteome study of DIAN participants found 124 proteins that went up (red) and one that went down (blue) as symptoms approached. A42, p-tau, and tau shown as controls. [Courtesy of Shen et al., medRxiv.]
The DIAN study follows hundreds of families with inherited AD. The cohort includes people who carry presenilin and APP mutations, as well as their unaffected relatives. Previous studies in this cohort had identified proteome changes early in disease. For example, more than a decade ago, a study of 14 mutation carriers found 56 proteins up- or downregulated in the CSF, while a recent one in 22 mutation carriers found 66 (Jan 2012 news; Jul 2023 news). A larger study from Erik Johnson and colleagues at Emory University analyzed CSF samples from 286 mutation carriers and 184 noncarriers in DIAN, but examined only 59 proteins, linking 33 of them to the disease (Aug 2023 news). There had not yet been a large, unbiased proteomics study in this population.
For that, Cruchaga and colleagues assembled a large international team that included Johnson and colleagues. They also analyzed samples from the DIAN cohort, now comprising 291 carriers and 185 noncarriers. The participants’ mean age was 40, and about two-thirds of the FAD mutation carriers were asymptomatic. On average, participants were seven years shy of their estimated year of symptom onset (EYO). First author Yuanyuan Shen used the SomaScan commercial platform, which detects proteins that bind short oligonucleotides known as aptamers, to measure 6,163 proteins in a single CSF sample from each participant (Candia et al., 2017). By correlating protein abundance with EYO in these cross-sectional samples, Shen and colleagues estimated how protein levels changed as the volunteers aged.
This analysis turned up 125 proteins with divergent trajectories in mutation carriers compared with noncarriers. All but one of them, the synaptic protein NPTX2, ticked up in carriers over time. They included known AD proteins, such as NfL and neuregulin, but many had not been linked to AD previously. Overall, the set of 125 included extracellular matrix proteins SMOC2 and SLIT2, metabolic proteins such as PEBP1 and GPI, and cytoskeleton-related proteins such as STMN2 and PDLIM4. The findings jibed with previous DIAN studies. Of the 125 proteins, 24 had been examined in the earlier work, and all of these exhibited similar changes across studies.
Proteins that changed in coordinated fashion fell into several groups, or modules. In one, proteins started diverging from controls 13 years before EYO. This module consisted of 21 mostly neuronal proteins that were involved in NMDA synaptic signaling, cellular stress, and mitochondrial damage. In another module, proteins diverged 10 years before EYO. This set contained 24 neuronal and endothelial proteins involved in apoptosis, neuronal death, and immune response. In a third module, proteins changed from control levels about seven years before EYO. This module had 47 mostly microglial, astrocyte, and innate immune proteins.
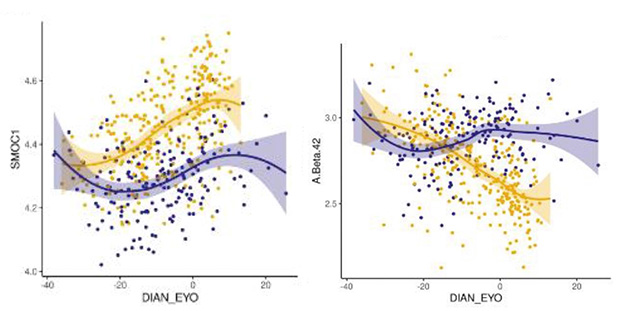
Earlier Marker. Extracellular matrix protein SMOC1 (left) begins to change 30 years prior to symptom onset in DIAN mutation carriers (gold) versus noncarriers (purple), beating Aβ42 (right) by 15 years. [Courtesy of Shen et al., medRxiv.]
Of particular interest was the ECM protein SMOC1, which rises 30 years before EYO and has turned up in several other proteome studies (Aug 2019 news; Feb 2022 news; Sep 2023 news). Betty Tijms and Lisa Vermunt at Amsterdam University Medical Center found this striking. “This indicates again that these SMOC proteins may be involved very early in the disease, and that we need to understand more about the processes related to this protein,” they wrote to Alzforum (comment below). Drummond agreed. “SMOC1 appears to be one of the most consistent fluid biomarkers of AD,” she wrote.
Using machine learning, Shen and colleagues identified nine proteins of the 125 that best identified people who carried an AD mutation. These were SMOC1, SMOC2, NPTX2, CAND1, PPP3CA, PRKCG, TMOD3, TREM1, and UBE2N. Together, the set nailed mutation status with an AUC of 0.89. This was better than the performance of p-tau181 and the p-tau/Aβ42 ratio, which both had an AUC of 0.80 in this cohort. However, p-tau and Aβ42 were better than the set of nine proteins at distinguishing symptomatic from presymptomatic carriers, consistent with the new set detecting very early changes, before symptoms appear.
“It is good to see replication of earlier findings from the Johnson study, and it is encouraging that both studies are detecting reproducible changes in CSF years before cognitive symptoms. This means the promise of early stage diagnostics could be realized,” noted Becky Carlyle at the University of Oxford, U.K.
What about biomarkers in plasma, which is easier to collect? These might be entirely different. Shen and colleagues found a much smaller set of 10 proteins that went up or down in ADAD plasma, with only SMOC1 in common with the CSF markers. Keenan Walker at the National Institute on Aging, Baltimore, noted that in sporadic AD, there is a large overlap between CSF and blood markers. “The findings suggest that, unlike late-life dementia syndromes, the proteomic signature for autosomal dominant AD is largely restricted to the CSF. By extension, this supports the idea that peripheral (non-CNS) factors play a larger role in determining risk for sporadic AD and related dementias emerging during late life,” he wrote to Alzforum (comment below).
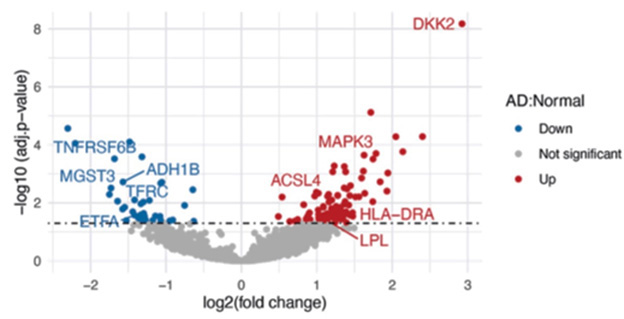
Plasma Proteins. Analysis of plasma from sporadic AD patients turned up a different set of elevated (red) and suppressed (blue) proteins. [Courtesy of Lacar et al., bioRxiv.]
Late-Onset AD Shows Up in Blood
In the second paper, Siddiqui and colleagues focused on the plasma signature of sporadic AD, and indeed found it was much more pronounced than in familial AD. Joint first authors Benjamin Lacar, Shadi Ferdosi, and Amir Alavi, all at Seer, used Proteograph technology, which detects proteins with nanoparticles, to measure more than 4,000 proteins in blood. Samples came from 1,006 participants in a longitudinal cohort study at the Massachusetts Alzheimer’s Disease Research Center. In this group, 379 people had AD, 387 other dementias, and 240 were cognitively healthy. The study included an average of two samples per participant.
Comparing AD to control samples, the authors identified 100 proteins that were more abundant in the former, and 38 that were less. Some of these, such as the kinase MAPK3, synaptic protein ACLS4, and apoptosis suppressor ADH1B, were known to have roles in AD. Others had not been directly tied to the disease before, but could help elucidate its mechanisms. For example, the most elevated protein, DKK2, inhibits the Wnt signaling pathway, which is known to be suppressed in AD. Metabolic protein MGST3 was down in AD patients, and has previously been associated with lower hippocampal size. Nonetheless, these plasma proteins did not perform as well as plasma p-tau181 in identifying AD patients, the authors noted.
The proteins might be more useful in predicting cognitive decline, with a set of eight associated with slippage on the CDR. Higher levels of BLVRB, CRISPLD2, CLNS1A, PRPS1, OXSR1, SELENBP1, SMYD5, and lower levels of GOLPH3, were linked to greater decline. “While some of the pathways and proteins identified are known to be involved with AD and related dementias, many are not and may point to novel biology,” the authors wrote. They believe the proteins predictive of decline could be used to aid treatment decisions in patients by flagging those most at risk.—Madolyn Bowman Rogers
References
News Citations
- Brain Proteins in Flux a Decade Before Familial AD Begins
- Proteomics Discerns Sporadic from Familial Alzheimer’s Disease
- Proteins in Biofluids Foreshadow Dementia by 30 Years
- Proteomics Uncovers Potential Markers, Subtypes of Alzheimer’s
- Proteomics Highlight Alzheimer’s Changes in Matrisome, MAPK Signaling
- CSF Proteomic Panel Better Predicts Decline Than Do Classic AD Biomarkers
Paper Citations
- Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, Huang J, Stuccio S, Zingone A, Ryan BM, Tsang JS, Biancotto A. Assessment of Variability in the SOMAscan Assay. Sci Rep. 2017 Oct 27;7(1):14248. PubMed.
Further Reading
Primary Papers
- Shen Y, Ali M, Liu M, Timsina J, Wang C, Do A, Western D, Gorijala P, Budde J, Liu H, Gordon B, Joseph-Mathurin N, Perrin RJ, McDade E, Morris JC, Llibre-Guerra JJ, Bateman RJ, Maschi D, Wyss-Coray T, Pastor P, Renton AE, Surace EI, Johnson EC, Alvarez I, Levin J, Ringman JM, Levey AI, Allegri RF, Seyfried N, Day GS, Wu Q, Fernandez MV, Ibanez L, Sung Y, Cruchaga C. Systematic proteomics in Autosomal dominant Alzheimer's disease reveals decades-early changes of CSF proteins in neuronal death, and immune pathways. 2024 Jan 13 10.1101/2024.01.12.24301242 (version 1) medRxiv.
- Lacar B, Ferdosi S, Alavi A, Stukalov A, Venkataraman GR, deGeus M, Dodge H, Wu C-Y, Kivisakk P, Das S, Guturu H, Hyman B, Batzoglou S, Arnold SE, Siddiqui A. Identification of Novel Biomarkers for Alzheimer's Disease and Related Dementias Using Unbiased Plasma Proteomics. 2024 Jan 08 10.1101/2024.01.05.574446 (version 1) bioRxiv.
Annotate
To make an annotation you must Login or Register.

Comments
University of Sydney
This is a nice study that extends the exciting findings reported in Johnson et al., 2023, by using an alternative proteomics approach—Somascan in comparison to the previously used SRM-MS—to quantify many more protein changes in the CSF in the same cohort of ADAD mutation carriers. Comparison between the studies showed many similarities, but still highlighted some discrepancies, which is not unexpected when using different proteomics approaches. Looking forward, additional studies, using different patient populations or different experimental approaches, will ultimately help the field refine which of these protein changes are robust, akin to what is now emerging with regards to AD-associated protein changes in the brain (Askenazi et al., 2023).
It is exciting that studies such as this one and Johnson et al. provide concrete evidence that AD-associated protein dysfunction (beyond Aβ and tau) are occurring up to 30 years prior to symptom onset. Of the proteins altered very early in disease, SMOC1 stands out. This study provides yet another example showing that SMOC1 appears to be one of the most consistent biofluid biomarkers of AD. Additionally, SMOC1 was the only protein change observed in both CSF and plasma samples in this study, providing additional evidence that it could also be an excellent plasma biomarker of AD. More research is needed to explore the role of SMOC1 in the brain and its link to AD. While the mechanism is unknown, we, and others, have shown that SMOC1 is highly enriched in amyloid plaques, suggesting it could have an important role in amyloid pathology (Drummond et al., 2022). Similarly, two other proteins, SLIT2 and SPON1, that increased in ADRD CSF more than 17 years prior to symptom onset are also highly enriched in amyloid plaques. SMOC2, altered ~ 22 years prior to symptom onset, was recently suggested to be associated with Aβ specifically in CAA (Wojtas et al., 2024). Together these results highlight understudied amyloid-associated proteins that should be examined in more detail in future studies.
Another interesting observation in this study was that while there were many similarities between ADAD and sporadic AD, some proteins were significantly altered in opposite directions in the CSF. This shows how important it is to consider potential divergent mechanisms between sporadic AD and ADAD when interpreting results, not only for studies using human tissue, but also when using animal models expressing ADAD mutations. Divergent protein changes were not common, since there were far more similarities between ADRD and sAD than not, but this caveat is still worth heeding.
References:
Askenazi M, Kavanagh T, Pires G, Ueberheide B, Wisniewski T, Drummond E. Compilation of reported protein changes in the brain in Alzheimer's disease. Nat Commun. 2023 Jul 25;14(1):4466. PubMed.
Drummond E, Kavanagh T, Pires G, Marta-Ariza M, Kanshin E, Nayak S, Faustin A, Berdah V, Ueberheide B, Wisniewski T. The amyloid plaque proteome in early onset Alzheimer's disease and Down syndrome. Acta Neuropathol Commun. 2022 Apr 13;10(1):53. PubMed.
Johnson EC, Bian S, Haque RU, Carter EK, Watson CM, Gordon BA, Ping L, Duong DM, Epstein MP, McDade E, Barthélemy NR, Karch CM, Xiong C, Cruchaga C, Perrin RJ, Wingo AP, Wingo TS, Chhatwal JP, Day GS, Noble JM, Berman SB, Martins R, Graff-Radford NR, Schofield PR, Ikeuchi T, Mori H, Levin J, Farlow M, Lah JJ, Haass C, Jucker M, Morris JC, Benzinger TL, Roberts BR, Bateman RJ, Fagan AM, Seyfried NT, Levey AI, Dominantly Inherited Alzheimer Network. Cerebrospinal fluid proteomics define the natural history of autosomal dominant Alzheimer's disease. Nat Med. 2023 Aug;29(8):1979-1988. Epub 2023 Aug 7 PubMed.
Wojtas AM, Dammer EB, Guo Q, Ping L, Shantaraman A, Duong DM, Yin L, Fox EJ, Seifar F, Lee EB, Johnson EC, Lah JJ, Levey AI, Levites Y, Rangaraju S, Golde TE, Seyfried NT. Proteomic Changes in the Human Cerebrovasculature in Alzheimer's Disease and Related Tauopathies Linked to Peripheral Biomarkers in Plasma and Cerebrospinal Fluid. 2024 Jan 11 10.1101/2024.01.10.24301099 (version 1) medRxiv.
Amsterdam UMC, loc. VUmc
Amsterdam UMC, VU University
This study by Shen et al. focusses on DIAN CSF SomaLogic 7k analyses and makes use of the estimated years of onset (EYO) concept to better understand disease processes that are associated with AD pathogenesis and their involvement over time.
It is particularly striking that with this platform, like the mass spec measurement in the same cohort, SMOC1 pops up as a very early marker of ADAD (Johnson et al., 2023). The runner-up strong effect was SMOC2, which was also picked up by Olink in our study (van de Ende, 2023). This indicates again that these SMOC proteins may be involved very early in the disease, and that we need to understand more about the processes that are related to them.
References:
Johnson EC, Bian S, Haque RU, Carter EK, Watson CM, Gordon BA, Ping L, Duong DM, Epstein MP, McDade E, Barthélemy NR, Karch CM, Xiong C, Cruchaga C, Perrin RJ, Wingo AP, Wingo TS, Chhatwal JP, Day GS, Noble JM, Berman SB, Martins R, Graff-Radford NR, Schofield PR, Ikeuchi T, Mori H, Levin J, Farlow M, Lah JJ, Haass C, Jucker M, Morris JC, Benzinger TL, Roberts BR, Bateman RJ, Fagan AM, Seyfried NT, Levey AI, Dominantly Inherited Alzheimer Network. Cerebrospinal fluid proteomics define the natural history of autosomal dominant Alzheimer's disease. Nat Med. 2023 Aug;29(8):1979-1988. Epub 2023 Aug 7 PubMed.
van der Ende EL, In 't Veld SG, Hanskamp I, van der Lee S, Dijkstra JI, Hok-A-Hin YS, Blujdea ER, van Swieten JC, Irwin DJ, Chen-Plotkin A, Hu WT, Lemstra AW, Pijnenburg YA, van der Flier WM, Del Campo M, Teunissen CE, Vermunt L. CSF proteomics in autosomal dominant Alzheimer's disease highlights parallels with sporadic disease. Brain. 2023 Nov 2;146(11):4495-4507. PubMed.
National Institute on Aging
Shen and colleagues applied the high-throughput SomaScan proteomic platform to measure more than 6,000 unique proteins in the CSF and plasma of participants with autosomal-dominant Alzheimer’s disease (ADAD) in the DIAN study, with the stated goal of informing disease biology and identifying biomarkers that may be used for treatment monitoring. This highly informative study adds an additional dimension to a well-characterized ADAD cohort in a manner that enhances our understand of the evolving disease biology in the decades leading up to dementia onset.
Especially revealing was the authors’ use of the estimated years of onset (EYO) to define protein-specific pseudo-trajectories using cross-sectional data. This clever approach provides a unique opportunity to determine when changes in specific proteins and specific protein groups begin to occur among mutation carriers. Of particular interest are the 12 proteins that changed in CSF before changes were observed for the core AD biomarkers (p-tau, total tau, and Aβ42). While changes to CSF p-tau, total tau, and Aβ42 occurred between 11 and 17 years before dementia onset, SMOC1 and SMOC2, two extracellular matrix proteins, were estimated to show deviations in CSF among mutation carriers approximately 30 years before dementia onset. These findings are supportive of other studies which show that proteins involved in extracellular matrix (ECM) organization and degradation are differentially expressed well before dementia onset in ADAD and sporadic AD (Johnson et al. 2023; Walker et al. 2023). These matrisome proteins have been hypothesized to interact with Aβ aggregates very early in the disease process.
These findings suggest that SMOC1 and SMOC2, and perhaps other matrisome proteins, may serve as the earliest biomarkers of AD pathology. Whether these proteins also show changes 30 years before dementia onset in CSF or plasma of individuals who develop sporadic AD remains to be seen.
One of the findings I found most interesting was the difference in number of proteins associated with ADAD in CSF as compared to plasma, and what this may tell us about the pathobiology of ADAD, as compared to sporadic AD. Given the direct contact of CSF with brain tissue, we would anticipate that the proteomic signature of ADAD is stronger in CSF that it is in plasma. That said, recent studies suggest that there is a fairly strong plasma proteomic signature associated with sporadic AD and all-cause dementia (ref Walker et al., 2021, 2023; Jiang et al., 2022). However, this does not appear to be the case for ADAD, as the authors found that only 9 of 6,022 plasma proteins were associated with ADAD at a false discovery rate p-value of less than0.05. In contrast, a comparable CSF analyses yielded 246 differentially expressed proteins in a smaller sample using the more stringent Bonferroni-corrected threshold. The findings suggest that, unlike late-life dementia syndromes, the proteomic signature for ADAD is largely restricted to the CSF. By extension, this supports the idea that peripheral (non-CNS) factors—and in some cases, the plasma proteins themselves—play a larger role in determining risk for sporadic AD and related dementias emerging during late-life.
Lastly, I’ll note that the plasma proteins found to be differentially expressed in ADAD are enriched for presynaptic function, and include synaptic proteins such as CPLX1 and CPLX2, which show altered levels in plasma decades ahead of symptom onset in non-ADAD dementia (ref Walker et al., 2023).
References:
Jiang Y, Zhou X, Ip FC, Chan P, Chen Y, Lai NC, Cheung K, Lo RM, Tong EP, Wong BW, Chan AL, Mok VC, Kwok TC, Mok KY, Hardy J, Zetterberg H, Fu AK, Ip NY. Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer's disease screening and staging. Alzheimers Dement. 2021 May 25; PubMed.
Johnson EC, Bian S, Haque RU, Carter EK, Watson CM, Gordon BA, Ping L, Duong DM, Epstein MP, McDade E, Barthélemy NR, Karch CM, Xiong C, Cruchaga C, Perrin RJ, Wingo AP, Wingo TS, Chhatwal JP, Day GS, Noble JM, Berman SB, Martins R, Graff-Radford NR, Schofield PR, Ikeuchi T, Mori H, Levin J, Farlow M, Lah JJ, Haass C, Jucker M, Morris JC, Benzinger TL, Roberts BR, Bateman RJ, Fagan AM, Seyfried NT, Levey AI, Dominantly Inherited Alzheimer Network. Cerebrospinal fluid proteomics define the natural history of autosomal dominant Alzheimer's disease. Nat Med. 2023 Aug;29(8):1979-1988. Epub 2023 Aug 7 PubMed.
Walker KA, Chen J, Shi L, Yang Y, Fornage M, Zhou L, Schlosser P, Surapaneni A, Grams ME, Duggan MR, Peng Z, Gomez GT, Tin A, Hoogeveen RC, Sullivan KJ, Ganz P, Lindbohm JV, Kivimaki M, Nevado-Holgado AJ, Buckley N, Gottesman RF, Mosley TH, Boerwinkle E, Ballantyne CM, Coresh J. Proteomics analysis of plasma from middle-aged adults identifies protein markers of dementia risk in later life. Sci Transl Med. 2023 Jul 19;15(705):eadf5681. PubMed.
Walker KA, Chen J, Zhang J, Fornage M, Yang Y, Zhou L, Grams ME, Tin A, Daya N, Hoogeveen RC, Wu A, Sullivan KJ, Ganz P, Zeger SL, Gudmundsson EF, Emilsson V, Launer LJ, Jennings LL, Gudnason V, Chatterjee N, Gottesman RF, Mosley TH, Boerwinkle E, Ballantyne CM, Coresh J. Large-scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nat Aging. 2021 May;1(5):473-489. Epub 2021 May 14 PubMed.
Washington University School of Medicine
Adapted from a LinkedIn post.
In order to fully understand the biology of Alzheimer’s disease, to create new predictive models, and to identify novel causal and druggable targets, we need large-scale, unbiased omic analyses. For the last three years, my lab has generated the largest and most detailed proteomic atlas in brain, CSF, and plasma from Alzheimer’s, Parkinson’s disease, and other neurodegenerative diseases.
One example of how this data will lead to a better understanding of AD is Wang et al., 2024. In this paper, we integrated genetic and proteomic data to identify additional genes and proteins that are part of the TREM2 pathway. By performing multi-ethnic mapping, we were able to identify the two functional variants on the MS4A locus. On top of that, we identified and functionally characterized the two novel loci TGFB2 and Nectin2. These findings were possible by analyzing large human proteomic datasets, as it is unclear whether the current cell and animal models would have captured these new genes and protein-protein interactions.
Building on that, we generated the largest CSF proteomic dataset in terms of samples (more than 3,000) and proteins (more than 7,000) from sporadic (Cruchaga et al., 2024) and autosomal-dominant AD (Shen et al., 204).
In these two preprints, we identified and replicated more than 2,000 proteins associated with clinical and biomarker status in sporadic AD and more than 120 for autosomal-dominant AD. We later leveraged those proteins to create novel predictive models as well as to identify pathways associated with disease.
In these two studies we were able to generate, replicate, and validate novel predictive models that include a small number of proteins that predict AD with extremely high power. This is important as those novel predictive models are Aβ- and tau-independent. As current therapies are targeting Aβ and tau, those proteins cannot be used as biomarkers but for target engagement. Our studies present novel biomarkers that could be used to track disease status in individuals taking new therapies.
In our sporadic AD study, because of the large number of individuals and proteins, we were able to demonstrate that the proteins associated with AD follow four unique trajectories capturing different biological processes, including 1) death of glutamatergic and dopaminergic neurons, 2) activation of immune response and endolysosomal dysfunction, 3) brain plasticity and mechanism to compensate for AD-related pathology, and 4) proteins involved in cell-to-cell crosstalk including microglia and astrocytes.
Changes in the Aβ and hyperphosphorylated tau proteins in brain and cerebrospinal fluid (CSF) precede AD symptoms, making the CSF proteome a potential avenue to understand the pathophysiology and facilitate reliable diagnostics and therapies. While CSF and plasma Aβ and tau levels are considered the gold standard biomarkers for AD, additional models are needed as most of the current disease-modifying therapies in clinical trials target Aβ and/or tau with a limited outcome. In order to identify novel Aβ- and tau-independent proteomics biomarkers for AD, we measured and analyzed the protein expression levels of 7,029 analytes in CSF of 2,286 participants from multicenter cohorts. We employed a three-stage analytical approach (discovery, replication, and meta-analysis) to identify robust AD CSF proteomic alterations. Machine learning was implemented to create and validate highly accurate and replicable 11-protein model that predict classical AD biomarker positivity and clinical status. The predicted model is AD-specific as it does not perform well for other neurodegenerative disorders (e.g., Parkinson's disease, frontotemporal dementia, and dementia with Lewy bodies), and it can also identify people who will convert to AD as well as those AD cases with faster progression.
References:
Wang L, Nykänen NP, Western D, Gorijala P, Timsina J, Li F, Wang Z, Ali M, Yang C, Liu M, Brock W, Marquié M, Boada M, Alvarez I, Aguilar M, Pastor P, Ruiz A, Puerta R, Orellana A, Rutledge J, Oh H, Greicius MD, Le Guen Y, Perrin RJ, Wyss-Coray T, Jefferson A, Hohman TJ, Graff-Radford N, Mori H, Goate A, Levin J, Sung YJ, Cruchaga C. Proteo-genomics of soluble TREM2 in cerebrospinal fluid provides novel insights and identifies novel modulators for Alzheimer's disease. Mol Neurodegener. 2024 Jan 3;19(1):1. PubMed.
Cruchaga C, Ali M, Shen et al Y. Multi-cohort cerebrospinal fluid proteomics identifies robust molecular signatures for asymptomatic and symptomatic Alzheimer’s disease. https://doi.org/10.21203/rs.3.rs-3631708/v1 Research Square
Shen Y, Ali M, Liu M, Timsina J, Wang C, Do A, Western D, Gorijala P, Budde J, Liu H, Gordon B, Joseph-Mathurin N, Perrin RJ, McDade E, Morris JC, Llibre-Guerra JJ, Bateman RJ, Maschi D, Wyss-Coray T, Pastor P, Renton AE, Surace EI, Johnson EC, Alvarez I, Levin J, Ringman JM, Levey AI, Allegri RF, Seyfried N, Day GS, Wu Q, Fernandez MV, Ibanez L, Sung Y, Cruchaga C. Systematic proteomics in Autosomal dominant Alzheimer's disease reveals decades-early changes of CSF proteins in neuronal death, and immune pathways. 2024 Jan 13 10.1101/2024.01.12.24301242 (version 1) medRxiv.
Make a Comment
To make a comment you must login or register.