Research Brief: Lucky Seven—Htt N-Terminal Structure Solved
Quick Links
Like preschool teachers trying to arrange squirmy youngsters into a straight line, structural biologists spend days, even years, taming the unruly—in their case, coaxing fickle proteins to form crystals that reveal insight into three-dimensional structure. Through crystallography studies that experts describe as “heroic,” Mee Whi Kim and colleagues at the University of Texas Southwestern Medical Center, Dallas, have solved the secondary structure of huntingtin, the protein at the root of Huntington disease. In this month’s issue of Structure, the scientists report that huntingtin adopts a surprising variety of conformations in its N-terminal polyglutamine region, which is mutated in HD. This structural flexibility may not only explain why it has been so hard to crystallize the huntingtin protein, but also raises questions about the function of the polyglutamine stretch and how certain conformations may underlie toxicity. For their part, tau and alpha-synuclein, two other proteins blamed for driving neurodegenerative disease, are still resisting crystallographers’ efforts to visualize their 3D structure.
Scientists have toiled well over a decade trying to determine the structure of huntingtin, one of nine polyglutamine (polyQ)-containing proteins known to cause neurodegenerative disease when misfolded forms of the molecule aggregate in the brain. The current study unveils the first crystal structure of a polyQ-expanded protein (>Q10) in its native context. Seven was the magic number in this technological tour de force. “Because the structure itself is not well formed, it was very difficult to make the crystals, and the x-ray diffraction was very complicated,” Kim told ARF. “That’s why it took seven years—five to make the crystals, two to solve the structure.” Analyzing diffraction from 30 crystals, the researchers obtained structures for seven—each with the polyglutamine region in a different conformation.
The trick to getting huntingtin to crystallize was recognizing that despite the unpredictability of the polyglutamine stretch, the structures of the flanking N-terminal and polyproline regions remain fairly stable. Kim and colleagues did not crystallize the entire huntingtin protein. Rather, they expressed the first exon of 17-polyQ huntingtin in bacteria, using maltose binding protein as an N-terminal tag and a 19-amino acid tag at the C-terminal end. “Between these two contacts, I could capture my protein and get it to stabilize,” Kim said.
Consistent with predictions from prior studies, the region immediately upstream of the multi-conformational polyQ sequence invariably appeared as an alpha-helix, while the downstream region formed a classic proline helix in straight or kinked configuration (see PP1 and PP2 in image below).
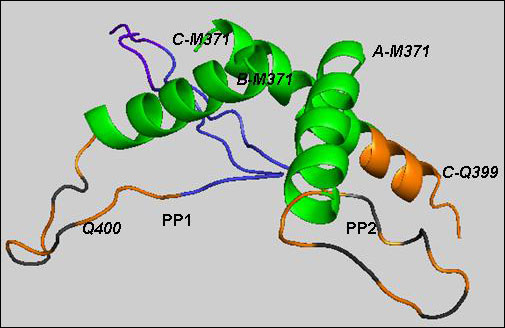
Structure of Htt17Q-EX1 trimer
Htt17Q-EX1 molecules (A, B, C) consist of a-helices and PP1 or PP2 (alternative conformations), connected by a random coil region. Htt17Q-EX1 begins with the amino-terminal a-helix from Met 371 to Phe387 (green). The following poly17Q region (orange) is shown as a-helix and a loop. PP1 and PP2 contain extended polyQ loop (orange), poly11P (Blue) and polyP/Q (purple) regions. The orange cartoon with black stripe is the random coil region of polyQ. Image courtesy of Mee Whi Kim.
In an accompanying commentary, Paul Muchowski of the University of California, San Francisco, speculates on possible functional roles for the conformational flexibility of huntingtin’s polyQ region. About half of the 66 human proteins that contain at least five glutamine repeats demonstrate considerable length polymorphism in the polyQ stretch. To Muchowski, this suggests that the ability to assume a variety of conformations might be essential for normal proteins that need to function within a wide range of polyQ lengths. He also notes that polyQ proteins are overrepresented in transcription-related processes, raising the possibility that conformational flexibility is important for these activities.
Another looming question is the N-terminal structure of pathological (Q36 or greater) huntingtin protein. The current study examined 17-polyQ huntingtin, which falls within the range of polyQ lengths in the normal population. “We still don't know the toxic conformation,” Kim said. “We have to work on proteins with longer glutamine stretches to see the conformation change there.” Based on the published data, she and colleagues propose that huntingtin’s pathogenic polyQ expansion “increases the length of the random coil, which promotes aggregation and facilitates abnormal interactions with other proteins in cells.”
Researchers are beginning to study polyQ stretches and other short tandem repeats on a whole-genome basis, as reported in a separate study published in this week’s PNAS Early Edition. Michael Molla of Boston University, Massachusetts, and colleagues catalogued all triplet repeats and their flanking regions. These authors propose that such data can be used to eventually design a cost-effective whole-genome assay to determine disease risk or to guide personalized genomic strategies relevant to drug response.—Esther Landhuis
References
No Available References
Further Reading
Papers
- Legleiter J, Lotz GP, Miller J, Ko J, Ng C, Williams GL, Finkbeiner S, Patterson PH, Muchowski PJ. Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant huntingtin fragment. J Biol Chem. 2009 Aug 7;284(32):21647-58. PubMed.
Primary Papers
- Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009 Sep 9;17(9):1205-12. PubMed.
- Miller J, Rutenber E, Muchowski PJ. Polyglutamine dances the conformational cha-cha-cha. Structure. 2009 Sep 9;17(9):1151-3. PubMed.
- Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):17095-100. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.