Tau Toggling Peptides: One Seeds Fibrils; the Other Dismantles Them
Quick Links
The accumulation of neurofibrillary tangles is the common thread among all tauopathies, yet each amasses fibrils with a distinct fold at their core. What forces goad tau into twisting into these specific folds in the first place, and once fibrils form, how can they be undone? Two studies chipped away at these complementary questions by studying peptides. One, led by David Eisenberg at the University of California, Los Angeles, and described in a bioRxiv preprint posted March 29, found that D-enantiomer peptides designed to interfere with tau fibrillization go one step further, completely dismantling existing fibrils extracted from an Alzheimer’s disease brain. Upon binding to AD-tau filaments, which are made of naturally occurring L-amino acids, these D-peptides twisted to conform to the fibril template. However, the match was not meant to be, with the strain of the D-fibril-L-fibril interaction ultimately causing both to fall apart.
- A batch of synthetic D-enantiomeric peptides untangle tau fibrils.
- These D-peptides form fibrils themselves, which pair with tau fibrils and disassemble them.
- A sequence between two microtubule binding domains of 4R-tau sparks misfolding.
- The motif incites prion-like propagation of 4R-tau fibrils.
In the second study, reported April 9 in Proceedings of the National Academy of Sciences, researchers led by Kenneth Kosik at the University of California, Santa Barbara, and Songi Han at Northwestern University in Chicago, leveraged a peptide sequence from within tau itself to understand more about the forces that drive seeded propagation. They found that a 19-residue peptide, corresponding to the junction between the second and third microtubule binding domains of tau, not only formed its own fibrils, but served as a potent seed to bring other 4R-tau molecules into the fold. A single mutation squelched the seeding activity of the peptide by obscuring its most amyloid-provoking sequence.
Together, the studies underscore mechanisms at the heart of tau seeding and propagation, and point the way to potential therapeutic strategies.
Both studies were informed by fibril structures found in different tauopathies (Oct 2021 news). As their name implies, 4R-tauopathies, such as progressive supranuclear palsy and corticobasal degeneration, are marked by tau fibrils spun of 4R tau, which harbors four microtubule binding repeat domains—R1-R4—the latter three forming the fibril core. In other tauopathies, such as Pick’s disease, fibrils are made of 3R tau, in which the second repeat is spliced out, leaving the remaining three-repeat domains to form fibril cores. For mixed 3R/4R tauopathies, such as AD, R3 and R4 domains contribute to the core. Within these repeat domains, researchers have identified key six-amino acid stretches— VQIINK in the R2 domain and VQIVYK in the R3 domain—that play a pivotal role in fibril formation. These sequences can form fibrils all on their own. However, these peptide fibrils are not capable of seeding other tau molecules to misfold.
Fibril-Busting Peptides
The amyloidogenic hexapeptides—also called PHF6— have long been in the crosshairs of scientists aiming to target tau. Previously, Eisenberg’s group used computational methods to design synthetic peptides targeting this sequence in the R3 domain. These capped the ends of growing tau fibrils, stunting their growth (Sievers et al., 2011). A six-residue D-enantiomer peptide, D-TLKIVW, was among the top cappers. Later, when the researchers attached this peptide to magnetic nanoparticles to aid its delivery into the brain, they were in for a surprise: the modification had somehow transformed the peptide from a mere fibril capper to a fibril destroyer. They determined that the nanoparticle itself was not responsible for the improved properties of the peptide. This left open the possibility that it was the seventh residue—a cysteine—that the researchers had added to the peptide to link it to the nanoparticle.
In their new study, first author Ke Hou and colleagues set out to understand what had transformed their D-peptides into fibril disassemblers. They started by generating a series of seven-residue, D-TLKIVWX peptides, with different residues at the end. Many of these heptapeptides triggered the disassembly of tau fibrils extracted from the AD brain, as viewed by transmission electron microscopy. This activity was most powerful when the seventh residue was a hydrophobic one, with isoleucine rounding out the most potent disaggregating peptides. When incubated with D-TLKIVWI for two days, AD tau fibrils disintegrated into smaller pieces, which were unable to seed tau fibrillization in biosensor cell lines.
In addition to the vestiges of tau fibrils, another entity emerged over time in the reactions. Much to the researchers’ astonishment, several of the D-TLKIVWX peptides formed fibrils themselves, and those that could not, including the original hexapeptide D-TLKIVW and D-TLKIVWP—also happened to be those that failed to disassemble tau fibrils. When the researchers took a closer look at these D-fibrils with atomic force microscopy and cryo-EM, they resolved right-twisted fibrils. All natural tau fibrils twist left. Some of the D-peptides had different structural polymorphs, but one thing they all had in common was a steric zipper motif, in which two b-sheets are snugly sandwiched together.
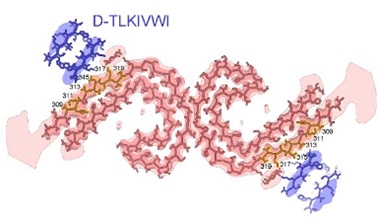
Takes One to Destroy One. Paired C-shaped protofilaments of tau (pink) are partnered with protofilaments of D-TLKIVWI. [Courtesy of Hou et al., bioRxiv, 2024.]
How might fibrillization of these peptides relate to their ability to disaggregate tau fibrils? To find out, the researchers cryogenically trapped D-peptides in the act of binding to AD tau fibrils. Under the electron microscope, they spotted extra densities paired with the outer surface of each C-shaped protofilament of tau. These cling-ons each represented a single unit of the D-peptide, folded into a steric zipper (image at right). Each of these peptide monomers stacked in each layer of the tau fibril, forming an associated D-peptide fibril.
Left to their own devices, these D-peptides fold into fibrils that twist to the right. However, when partnered with left-twisting tau, they tried to conform, adopting the uncomfortable position of twisting to the left—at least for a time. Eisenberg thinks that eventually the D-peptide fibrils might attempt to wriggle to the right to relieve the strain. This “strain relief” might be what ultimately leads to the undoing of both fibrils, he proposed (image below).
“We think it takes a fibril to destroy one,” Eisenberg said.
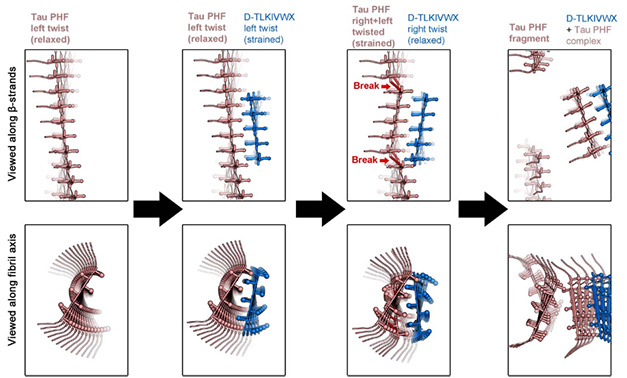
Destructive Relief. In the proposed strain relief mechanism, left-twisted tau fibrils bind D-peptides, which initially adopt tau’s left-handed twist to form paired fibrils. Structurally strained, D-peptide fibrils attempt to relax by twisting back to the right, causing both fibrils to fall apart. [Courtesy of Hou et al., bioRxiv, 2024.]
Interestingly, this stress, and its subsequent relief, would not have happened had the scientists used L-peptides, that naturally twist to the left, Eisenberg explained. He said D-peptides hold promise as therapeutics, noting that they come with the benefit of being more stable and avoiding immune clearance in the body.
Marc Diamond of UT Southwestern in Dallas called the paper an important conceptual advance. “[It] sheds important light on how one might engineer small molecules or biological agents to bind and ‘strain’ the association of monomer with filaments, leading to their disassembly,” he wrote. “The fact that these peptides do so as amyloid structures is even more interesting.” Kosik agreed. “I think it’s a beautiful paper,” he said. The fact that the peptide had to twist into a fibril to dismantle tau fibrils is quite intriguing, he added. Still, Diamond noted that the results were obtained in vitro and at high concentrations of peptides.
Eisenberg said that in a paper in press at Science Advances, his group reports the tau-busting activity of the D-peptides in vivo in mouse models of tauopathy.
Coaxing Tau to Tangle
Kosik and colleagues took a different approach, using the tau sequence itself to understand how tau fibrils form, and how to stop it. Focusing on 4R tau, first author Andrew Longhini and colleagues searched for the minimal stretch of the protein capable of the templated misfolding that sets off fibrillogenesis. As a guide, they used the structure of 4R tau fibrils derived from cryo-EM, which revealed a distinct hairpin turn formed by the R2 and R3 domains. Recently, his group reported that jR2R3, a 19-residue sequence that spans the R2-R3 junction, can form fibrils all on its own, and does so more readily if the FTD-linked, R2 domain mutation P301L is present (Vigers et al., 2023). For the new study, they asked whether this peptide was capable of corrupting naïve tau proteins into misfolding, and if so, how.
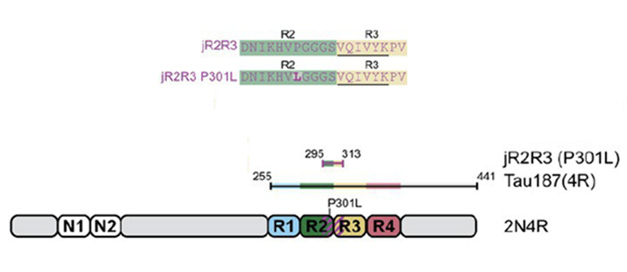
Fibrillogenic Peptides. The jR2R3 and jR3R3 P301L peptides (top) span the junction of R2 and R3 domains of tau (bottom). The Tau187 construct used in some experiments includes all four repeat domains and the C-terminus of tau. [Courtesy of Hou et al., PNAS, 2024]
What did they find? First, that both jR2R3 and jR2R3-P301L fibrils were capable of provoking other peptide monomers, as well as larger tau species such as full-length wild type or P301L tau, or the N-terminally truncated Tau187 fragment, into fibrillizing in a test tube. In cells, jR2R3-P301L fibrils robustly seeded aggregation of P301L tau monomers, which fibrillized and clumped into solid inclusions. JR2R3-P301L fibrils also seeded aggregation of wild-type tau, albeit much less efficiently. What’s more, insoluble material extracted from seeded cells robustly sparked fibril formation in other cells, a tenet of template-like propagation (image below). The seeding capacity of jR2R3-P301L was highly specific for 4R tau.
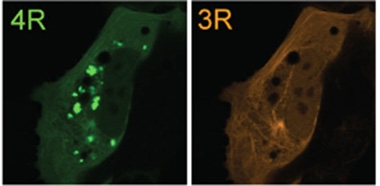
Seeding Specificity. Extracts of cells that had been seeded with jR2R3 fibrils seeded aggregation of 4R tau (green, inclusions) but not 3R tau (orange). [Courtesy of Longhini et al., PNAS, 2024.]
How did jR2R3-P301L accomplish its 4R-tau seeding feat? The researchers investigated by systematically mutating each of the four residues that distinguish the R2R3 junction of 4R tau from the corresponding R1R3 junction in 3R isoforms. Ultimately, they found that a single tweak—mutating residue 305 in jR2R3 from a serine to a lysine—completely abolished its seeding activity. The scientists found that while hairpin structures formed by the R1R3 peptide and S305L jR2R3 featured extensive intramolecular hydrogen bonds that kept the amyloidogenic PHF6 sequence buried in the peptide, R2R3 forged fewer of these internal contacts, leaving the PHF6 sequence exposed for templating.
The findings jibe with previous work. Diamond’s group identified seed competent species of tau monomers, reporting that their seeding capacity boiled down to the relative exposure of aggregation-prone PHF6 sequence (Aug 2017 conference news; Mirbaha et al., 2018). Collaborating with Diamond, Lukasz Joachimiak’s group at UT Southwestern reported that these sequences are relatively cloistered in 3R tau, and that disease-causing mutations such as P301L can open them up (Chen et al., 2019; Bali et al., 2023).
Kosik thinks these findings point to potential therapeutics. While a more challenging approach could use CRISPR to make the S305K switch in people at risk of a 4R tauopathy, other therapies already in preclinical development might also do the trick, he told Alzforum. For example, the tau specific Z70 nanobody—an antibody fragment that consists only of the antigen-snagging variable heavy chain domain—binds to the PHF6 sequence (May 2023 conference news; Mar 2024 conference news). Longhini and colleagues found that in cells expressing the nanobody, the amyloid-prone sequence was obscured, protecting naïve tau monomers from corruption by jR2R3-P301L fibrils.
In the same vein, Diamond and Joachimiak have generated potent, anti-seed antibodies that prevent exposure of tau’s aggregation-prone sequences (Hitt et al., 2023).—Jessica Shugart
References
News Citations
- Flock of New Folds Fills in Tauopathy Family Tree
- Monomeric Seeds and Oligomeric Clouds—Proteopathy News from AAIC
- New Arrows Aimed at Tau: Single-Domain Antibody, Peptibody, Vaccine
- Therapeutic Contenders Target Hard-to-Reach Pockets of Tau
Paper Citations
- Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Münch J, Baker D, Eisenberg D. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011 Jul 7;475(7354):96-100. PubMed.
- Vigers M, Lobo S, Najafi S, Dubose A, Tsay K, Ganguly P, Longhini AP, Jin Y, Buratto SK, Kosik KS, Shell MS, Shea J-E, Han S. Tau P301L mutation promotes core 4R tauopathy fibril fold through near-surface water structuring and conformational rearrangement. 2023 Nov 28 10.1101/2023.11.28.568818 (version 1) bioRxiv.
- Mirbaha H, Chen D, Morazova OA, Ruff KM, Sharma AM, Liu X, Goodarzi M, Pappu RV, Colby DW, Mirzaei H, Joachimiak LA, Diamond MI. Inert and seed-competent tau monomers suggest structural origins of aggregation. Elife. 2018 Jul 10;7 PubMed.
- Chen D, Drombosky KW, Hou Z, Sari L, Kashmer OM, Ryder BD, Perez VA, Woodard DR, Lin MM, Diamond MI, Joachimiak LA. Tau local structure shields an amyloid-forming motif and controls aggregation propensity. Nat Commun. 2019 Jun 7;10(1):2493. PubMed.
- Bali S, Singh R, Wydorski PM, Wosztyl A, Perez VA, Chen D, Rizo J, Joachimiak LA. Ensemble-based design of tau to inhibit aggregation while preserving biological activity. bioRxiv. 2023 Dec 14; PubMed.
- Hitt BD, Gupta A, Singh R, Yang T, Beaver JD, Shang P, White CL 3rd, Joachimiak LA, Diamond MI. Anti-tau antibodies targeting a conformation-dependent epitope selectively bind seeds. J Biol Chem. 2023 Nov;299(11):105252. Epub 2023 Sep 14 PubMed.
Further Reading
No Available Further Reading
Primary Papers
- Hou K, Ge P, Sawaya MR, Dolinsky JL, Yang Y, Jiang YX, Lutter L, Boyer DR, Cheng X, Pi J, Zhang J, Lu J, Yang S, Yu Z, Feigon J, Eisenberg DS. How short peptides can disassemble ultra-stable tau fibrils extracted from Alzheimers disease brain by a strain-relief mechanism. 2024 Mar 29 10.1101/2024.03.25.586668 (version 1) bioRxiv.
- Longhini AP, DuBose A, Lobo S, Vijayan V, Bai Y, Rivera EK, Sala-Jarque J, Nikitina A, Carrettiero DC, Unger MT, Sclafani OR, Fu V, Beckett ER, Vigers M, Buée L, Landrieu I, Shell S, Shea JE, Han S, Kosik KS. Precision proteoform design for 4R tau isoform selective templated aggregation. Proc Natl Acad Sci U S A. 2024 Apr 9;121(15):e2320456121. Epub 2024 Apr 3 PubMed.
Follow-On Reading
Papers
- Hou K, Pan H, Shahpasand-Kroner H, Hu C, Abskharon R, Seidler P, Mekkittikul M, Balbirnie M, Lantz C, Sawaya MR, Dolinsky JL, Jones M, Zuo X, Loo JA, Frautschy S, Cole G, Eisenberg DS. D-peptide-magnetic nanoparticles fragment tau fibrils and rescue behavioral deficits in a mouse model of Alzheimer's disease. Sci Adv. 2024 May 3;10(18):eadl2991. Epub 2024 May 1 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.