New Genetic Almanac Should Help Scientists Understand GWAS Hits
Quick Links
Genome-wide association studies point scientists toward chromosome loci that influence disease risk, but for the most part they yield ZIP codes rather than specific genetic addresses, many of them in non-coding regions. “GWAS have undoubtedly provided insight … but they do not necessarily tell us the important disease-linked genes,” said Mina Ryten of University College London. In the August 31 Nature Neuroscience online, Ryten and the U.K. Brain Expression Consortium describe a new resource to help do just that. They have created a database of expression-quantitative trait loci (eQTLs)—single-nucleotide polymorphisms (SNPs), either within or outside protein-coding genes, that somehow influence the expression of one or more genes. Covering 10 different regions of the brain, the Brain eQTL Almanac includes some that are important in neurodegeneration, such as the hippocampus and substantia nigra.
“Once people start rolling up their sleeves and digging around in the data to look at their favorite genes, we will see the value of this [database],” commented Alison Goate of Washington University in St. Louis, who was not involved in the study. Researchers may start with an SNP and look for the genes it affects, or start with a gene and use the data to build a network of eQTLs that regulate it and related genes, Ryten explained.
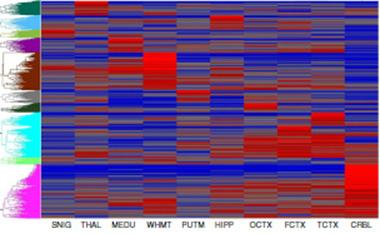
A chart of eQTL data. Each row represents an eQTL and each column one of 10 brain regions studied. Stronger or weaker eQTL activities are represented by red and blue, respectively. In each region, a cluster of certain eQTLs, indicated by the colored groups on the left, was more active (see text below). [Image courtesy of Ramasamy et al., Nature Neuroscience.]
Ryten and co-senior author Michael Weale of King’s College London led the work as part of a large collaboration with the U.K. Brain Expression Consortium and North American Brain Expression Consortium to analyze postmortem tissues from 134 neurologically healthy brain donors. They selected the 10 brain regions based on the collaborators’ interest in neurological disease. In addition to the hippocampus and substantia nigra, they looked at the cerebellar cortex, frontal cortex, inferior olivary nucleus, occipital cortex, putamen, temporal cortex, thalamus, and intralobular white matter. Ryten would have liked to test even more regions, but funding was limited. However, the authors continue to collect tissues to add to the database. First author Daniah Trabzuni of University College London isolated both DNA and RNA from those samples to determine SNP profiles and gene-expression levels.
To identify eQTLs, co-first author Adaikalavan Ramasamy, now at Oxford University in the United Kingdom, led a large and complex genome-wide association study (GWAS). Traditionally a GWAS looks for SNPs that are associated with a binary phenotype—disease or no disease. In this case, Ramasamy considered a series of continuous phenotypes, that is, the RNA levels of all genes in each tissue type. In addition, he correlated SNPs to splicing patterns. From this analysis, co-first author Sebastian Guelfi of UCL assembled the Brain eQTL Almanac database. One shortfall is that while eQTLs may occur far from their target gene—even on different chromosomes—the study lacked power to detect these distant modifiers, Ryten said. The authors limited their analysis to so-called cis-eQTLs, which are located within one megabase of the gene they influence.
The researchers found about 30,000 cis-eQTLs that regulated 8,573 genes in some fashion. Because they were interested in neurological disease, they compared their eQTLs to a list of 385 previously published SNPs from the U.S. National Human Genome Research Institute catalog. These 385 associate with brain-related traits such as Parkinson’s. About 17 percent of these SNPs appeared in the eQTL database, as well. About 23 percent of SNPs linked specifically to adult-onset neurological disorders also appeared in the eQTLs almanac. In this paper, the study authors examined just a few of those SNPs in detail, offering a glimpse into what their database can really do.
Sometimes the eQTL showed up within the gene it regulated. For example, a risk SNP for Alzheimer’s disease that lies within the complement receptor 1 (CR1) gene on chromosome 1 turned out to be an eQTL for the CR1 gene (see Jun 2010 news story). No big surprise.
More exciting to Ryten were eQTLs that pointed at the existence of novel disease genes at some distance from the SNP itself. There were several such cases: 61 out of 149 cis-eQTL signals for brain diseases were completely distinct from their target gene. For example, a chromosome 19 risk SNP for amyotrophic lateral sclerosis lies in an intron of the gene UNC13A, which regulates neurotransmitter release and seemed a shoo-in for an ALS risk gene (see Sep 2009 news story). However, the Brain eQTL Almanac indicates that the ALS SNP regulates not UNC13A, but a potassium channel called the potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 (KCNN1). KCNN1 controls neuron excitability.
Curiously, the eQTL correlated with KCNN1 expression only in the frontal cortex. The authors do not know how a frontal cortex eQTL could influence ALS risk. Perhaps the SNP is also an eQTL for KCNN1 in motor neurons of the spinal cord, suggested Ryten, who noted that spinal cord tissue was excluded from the analysis because it is hard to obtain. Based on their functions, either UNC13A or KCNN1 could be the real ALS risk factor, the authors wrote, but they argued that the eQTL data make a stronger case for KCNN1.
The authors also examined a Parkinson’s disease-related SNP (Pankratz et al., 2012). Because it sits in a non-coding region of chromosome 6 that is chock-full of genes, researchers have struggled to determine which gene it affects. In the eQTL database, that SNP points to the gene for major histocompatibility complex, class II, DQ alpha 2 (HLA-DQA2). Several studies have previously linked immune cells and inflammation to PD (see Apr 2009 news story; Feb 2009 news story; Jul 2008 news story).
In addition to investigating GWAS hits, scientists can use the almanac to understand the basic biology of gene regulation in the brain. For example, each brain tissue contained a cluster of eQTLs that were most influential (see image above). That means every brain region has its own particular way of regulating genes. This finding may help to explain why an inherited mutation, though it occurs in every cell in the body, can cause disease in a specific part of the brain. For example, Parkinson’s genes may be subject to specific regulation in the substantia nigra, making cells in that region more vulnerable. If substantiated, this approach might help scientists understand the frequently discussed problem of selective vulnerability in neurodegenerative disease.
With these examples, the study authors have “just scratched the surface,” said Goate. Philip De Jager of Harvard University, who did not participate in the work, agreed. “This study significantly extends the range of brain regions that have been profiled and examined to identify eQTLS,” he wrote in an email to Alzforum. “It will serve as an excellent resource for future investigations, particularly those involving neuropsychiatric diseases that have regionally specific effects” (see full comment below).—Amber Dance
References
News Citations
- Repeat Offenders—CLU, CR1, PICALM Hold Up in Association Studies
- Research Brief: Latest ALS GWAS Points to Loci on Chromosomes 9, 19
- Out-of-Control Inflammation in Parkinson's: It's Glia, Again
- Sleep On It—Astrocytes May Play Key Roles in PD, AD
- A PD Trifecta: Synuclein Gene Expression, Aging, and Inflammation
Paper Citations
- Pankratz N, Beecham GW, DeStefano AL, Dawson TM, Doheny KF, Factor SA, Hamza TH, Hung AY, Hyman BT, Ivinson AJ, Krainc D, Latourelle JC, Clark LN, Marder K, Martin ER, Mayeux R, Ross OA, Scherzer CR, Simon DK, Tanner C, Vance JM, Wszolek ZK, Zabetian CP, Myers RH, Payami H, Scott WK, Foroud T, . Meta-analysis of Parkinson's disease: identification of a novel locus, RIT2. Ann Neurol. 2012 Mar;71(3):370-84. PubMed.
External Citations
Further Reading
Papers
- Holton P, Ryten M, Nalls M, Trabzuni D, Weale ME, Hernandez D, Crehan H, Gibbs JR, Mayeux R, Haines JL, Farrer LA, Pericak-Vance MA, Schellenberg GD, , Ramirez-Restrepo M, Engel A, Myers AJ, Corneveaux JJ, Huentelman MJ, Dillman A, Cookson MR, Reiman EM, Singleton A, Hardy J, Guerreiro R, Apostolova LG, Arnold SE, Baldwin CT, Barber R, Barmada MM, Beach TG, Beecham GW, Beekly D, Bennett DA, Bigio EH, Bird TD, Blacker D, Boeve BF, Bowen JD, Boxer A, Burke JR, Buros J, Buxbaum JD, Cairns NJ, Cantwell LB, Cao C, Carlson CS, Carney RM, Carrasquillo MM, Carroll SL, Chui HC, Clark DG, Cotman CW, Crane PK, Crocco EA, Cruchaga C, Cummings JL, De Jager PL, Decarli C, Dekosky ST, Demirci FY, Diaz-Arrastia R, Dick M, Dickson DW, Duara R, Ellis WG, Ertekin-Taner N, Evans D, Faber KM, Fallon KB, Farlow MR, Ferris S, Foroud TM, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Goate AM, Graff-Radford NR, Green RC, Growdon JH, Hakonarson H, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jarvik GP, Jicha GA, Jin LW, Jun G, Kamboh MI, Karlawish J, Karydas A, Kauwe JS, Kaye JA, Kim R, Koo EH, Kowall NW, Kramer P, Kukull WA, Lah JJ, Larson EB, Levey AI, Lieberman AP, Lopez OL, Lunetta KL, Mack WJ, Marson DC, Martin ER, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Montine TJ, Morris JC, Naj AC, Nowotny P, Parisi JE, Peskind E, Petersen RC, Poon WW, Potter H, Quinn JF, Raj A, Rajbhandary RA, Raskind M, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rogaeva E, Rosenberg RN, Sano M, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Shelanski ML, Smith CD, Sonnen JA, Spina S, St George-Hyslop P, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Tsuang DW, Valladares O, Van Deerlin VM, Vardarajan BN, Vinters HV, Vonsattel JP, Wang LS, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Wright CB, Younkin SG. Initial assessment of the pathogenic mechanisms of the recently identified Alzheimer risk Loci. Ann Hum Genet. 2013 Mar;77(2):85-105. PubMed.
- Wang X, Chen Y, Lu L. Genetic regulatory network analysis for app based on genetical genomics approach. Exp Aging Res. 2010 Jan-Mar;36(1):79-93. PubMed.
- Myers AJ. AD gene 3-D: moving past single layer genetic information to map novel loci involved in Alzheimer's disease. J Alzheimers Dis. 2013;33 Suppl 1:S15-22. PubMed.
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, 't Hoen PA, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Völker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JB, Franke L. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013 Oct;45(10):1238-43. PubMed.
- Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, Gibbs JR, Ryten M, Arepalli S, Weale ME, Zonderman AB, Troncoso J, O'Brien R, Walker R, Smith C, Bandinelli S, Traynor BJ, Hardy J, Singleton AB, Cookson MR. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis. 2012 Jul;47(1):20-8. PubMed.
- Shen Q, Wang X, Chen Y, Xu L, Lu L. Expression QTL and regulatory network analysis of microtubule-associated protein tau gene. Parkinsonism Relat Disord. 2009 Aug;15(7):525-31. PubMed.
News
- Alzheimer's GWAS Hits Reflected in Monocyte Gene Expression
- Researchers Build on GWAS to Parse Genetic Players in AD and PD
- RNA Sequencing Helps Identify Functional Variants from GWAS
- Exome–Network Combination Uncovers New Disease Genes
- Pooled GWAS Reveals New Alzheimer’s Genes and Pathways
- Newly Mapped DNA Elements Help Interpret GWAS
- ENCODE Turns Human Genome From Sequence to Machine
Primary Papers
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, De T, UK Brain Expression Consortium, North American Brain Expression Consortium, Coin L, de Silva R, Cookson MR, Singleton AB, Hardy J, Ryten M, Weale ME, UK Brain Expression Consortium, North American Brain Expression Consortium. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014 Oct;17(10):1418-28. Epub 2014 Aug 31 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
Columbia University Irving Medical Scool
This is an interesting study that significantly extends the repertoire of genetic variations that influence gene expression. As has been shown in several recent studies, many genetic variations associated with disease susceptibility influence gene expression in cell- and context-specific manners. This study significantly extends the range of brain regions that have been profiled and examined to identify expression-quantitative trait loci (eQTLs); it will serve as an excellent resource for future investigations, particularly those involving neuropsychiatric disease that have regionally specific effects.
In addition, having profiled 10 brain regions in a substantial number of subjects, the authors enable robust comparisons across the regions to address how gene expression correlates with functional specificity, and they uncover interesting observations such as the co-variation of certain cis-eQTL signals in specific brain regions. These patterns suggest higher-order structure in gene expression that will refine our understanding of the brain transcriptome. Extending these types of studies to understand which cell types are driving the association signal is an important goal for future studies.
View all comments by Philip De JagerWashington University School of Medicine
Genome-wide associations studies (GWAS) have been extremely successful in identifying novel loci for complex traits, including Alzheimer’s disease (AD). The latest and largest GWAS for Alzheimer disease identified 19 genomic regions associated with risk (Lambert et al., 2013). Despite this success, we are still far from understanding the biological role of these associations. In some cases, it is not clear what gene or genetic variant is responsible for the association, and in other cases we do not know what is the functional mechanism driving the association.
Because some of those signals are not in the coding region but close to the gene, it has been speculated that those SNPs may affect overall gene-expression or splicing. Several studies have previously analyzed the association of gene expression with different genetic variants (Allen et al., 2012; Karch et al., 2012), and found that some of the GWAS signals also are associated with gene expression (eQTL).
In this study Ramasamy et al. moved a step forward and generated gene-expression data at the exon level in 10 different brain regions from 134 brains samples without known neurological disorders. They also generated GWAS data for all the individuals. They found a large number of eQTLs. Some of them were tissue- or region-specific, while others were not. Additional analyses identified pathways that may help us understand some of the biology of Alzheimer’s disease. Some of the eQTLs also overlap with genomic regions associated with complex traits such as Parkinson’s disease, ALS, lung cancer, or smoking and nicotine addiction
These are some examples of the results derived from this study. Much more results are presented in the paper with broad repercussion in complex traits. This project has generated so much data that additional results will be found with further analyses. More importantly all of this data is currently available on dbGAP or the GEO dataset, so any investigator can have access to the processed data or the raw data.
References:
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer's Disease Initiative (EADI), Genetic and Environmental Risk in Alzheimer's Disease, Alzheimer's Disease Genetic Consortium, Cohorts for Heart and Aging Research in Genomic Epidemiology, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O'Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, Wang J, Uitterlinden AG, Rivadeneira F, Koudstgaal PJ, Longstreth WT Jr, Becker JT, Kuller LH, Lumley T, Rice K, Garcia M, Aspelund T, Marksteiner JJ, Dal-Bianco P, Töglhofer AM, Freudenberger P, Ransmayr G, Benke T, Toeglhofer AM, Bressler J, Breteler MM, Fornage M, Hernández I, Rosende Roca M, Ana Mauleón M, Alegrat M, Ramírez-Lorca R, González-Perez A, Chapman J, Stretton A, Morgan A, Kehoe PG, Medway C, Lord J, Turton J, Hooper NM, Vardy E, Warren JD, Schott JM, Uphill J, Ryan N, Rossor M, Ben-Shlomo Y, Makrina D, Gkatzima O, Lupton M, Koutroumani M, Avramidou D, Germanou A, Jessen F, Riedel-Heller S, Dichgans M, Heun R, Kölsch H, Schürmann B, Herold C, Lacour A, Drichel D, Hoffman P, Kornhuber J, Gu W, Feulner T, van den Bussche H, Lawlor B, Lynch A, Mann D, Smith AD, Warden D, Wilcock G, Heuser I, Wiltgang J, Frölich L, Hüll M, Mayo K, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Singleton AB, Guerreiro R, Jöckel KH, Klopp N, Wichmann HE, Dickson DW, Graff-Radford NR, Ma L, Bisceglio G, Fisher E, Warner N, Pickering-Brown S. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013 Dec;45(12):1452-8. Epub 2013 Oct 27 PubMed.
View all comments by Carlos CruchagaMake a Comment
To make a comment you must login or register.