RNA Twist: C9ORF72 Intron Expansion Makes Aggregating Protein
Quick Links
Since the identification of the major amyotrophic lateral sclerosis and frontotemporal lobar degeneration gene C9ORF72 more than a year ago, scientists have been hunting for clues as to how it promotes disease. Being an expanded hexanucleotide repeat in an intron, i.e., non-coding region, the genetic defect causing ALS and FTLD should not affect the protein sequence. Ideas have ranged from loss of function due to RNA instability to toxic RNA inclusions. Now, in a big surprise, researchers led by Dieter Edbauer and Christian Haass at Ludwig-Maximilians University, Munich, Germany, report something entirely different. In today’s Science, they show that the expansion, in fact, gets translated. It produces aberrant dipeptide repeat (DPR) proteins. Not only that, but the authors also showed that neuronal inclusions present in mutation carriers contain these DPR proteins. The data help solve the mystery of these enigmatic inclusions and “link the genetic mutation to the predominant pathology,” Edbauer told Alzforum. What remains to be shown is whether these deposits actually drive the disease. Nonetheless, scientists hailed the discovery as a major advance.
“I think this is a great step forward in our understanding of the disease,” said Bryan Traynor at the National Institute on Aging in Bethesda, Maryland. He co-led one of the research groups that initially identified the C9ORF72 mutation, but was not involved in the current study. “It is a very exciting discovery. Of course, like all good science, this finding leads to more questions,” Traynor added. Stuart Pickering-Brown at the University of Manchester, U.K., wrote to Alzforum: “The next step is to clarify if the dipeptides are toxic, or just innocent byproducts of the expansion.”
Strengthening the findings, another group has independently arrived at the same conclusions. Researchers at the Mayo Clinic in Jacksonville, Florida, also found that the mysterious inclusions seen only in C9ORF72 mutation carriers contain DPR proteins made from the hexanucleotide expansion. That work will be published next week.
For years, researchers knew that an important genetic linkage to ALS and FTLD lurked on chromosome 9, but not until 2011 did two groups pin it down to a hexanucleotide repeat expansion in the first intron of the C9ORF72 gene (see ARF related news story). Most healthy people have 25 or fewer copies of the repeat, whereas mutation carriers can have 700 copies or more. The mutation explains roughly half of familial ALS cases and about a quarter of inherited FTLD, as well as some sporadic cases, making it the most important genetic risk factor for these disorders (see ARF related news story). So far, researchers have proposed two main theories for how the expansion could cause disease. One, it lowers levels of the C9ORF72 gene product, which may regulate membrane trafficking (see ARF related news story). Two, the mutant RNA forms intranuclear deposits that sequester key RNA-binding proteins and disrupt cell function (see ARF related news story).
Edbauer and colleagues considered a third possibility. Previously, researchers led by Laura Ranum, then at the University of Minnesota, Minneapolis, had shown that a CAG trinucleotide repeat expansion in the ataxin 8 gene permits the mutant RNA to form hairpin structures that stimulate aberrant translation in several reading frames. This repeat-associated, non-ATG-initiated (RAN) translation also occurs in another gene with a non-coding RNA expansion, namely dystrophia myotonica-protein kinase. In both cases, the resulting proteins form deposits characteristic of the movement disorders spinocerebellar ataxia type 8, and myotonic dystrophy type 1, respectively (see Zu et al., 2011). Edbauer wondered if the same mechanism could occur in C9ORF72 mutation carriers.
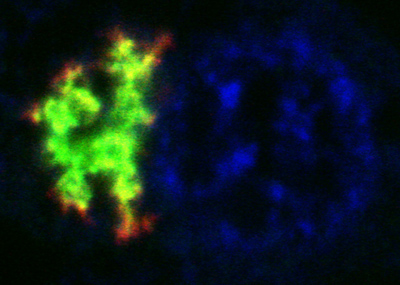
Joint first authors Kohji Mori and Shih-Ming Weng raised antibodies against the three possible DPRs that would be produced from the hexanucleotide expansion if it was translated in any of the three reading frames: glycine-alanine, glycine-proline, and glycine-arginine repeats. The authors detected all three DPRs in cerebellar extracts from postmortem brains of C9ORF72 mutation carriers, but none in brain samples taken from healthy controls or people with other forms of FTLD/ALS. The authors found poly-glycine-alanine to be the most abundant of the DPR proteins, although all three were present. Likewise, in brain slices from mutation carriers the antibodies labeled star-shaped cytoplasmic inclusions and a few dot-like intranuclear inclusions found in neurons (see image below).
DPR Proteins in Disease
Poly GA (red) co-localizes with p62 (green) in star-like inclusions in the hippocampus of a C9ORF72 mutation carrier. Image courtesy of Kohji Mori and Dieter Edbauer
Similar inclusions have previously been shown to be positive for the signaling protein p62 and negative for TDP-43, one of the primary pathological proteins in FTLD and ALS. The deposits occur in neurons of the hippocampus, cerebellum, and neocortex in people with the C9ORF72 mutation, but not in people with other forms of the disorders (see, e.g., Al-Sarraj et al., 2011; Mahoney et al., 2012; Bigio et al., 2012). It is not yet clear how these mostly cytoplasmic protein aggregates relate to the intranuclear RNA deposits also seen in carriers.
How do the new data relate to the findings that C9ORF72 mRNA levels are lower in carriers? Matching what other groups have found, the authors saw lowered levels of total mature C9ORF72 mRNA in the cerebellum of carriers compared to controls (see, e.g., Dejesus-Hernandez et al., 2011; Gijselinck et al., 2012). However, transcripts containing intron 1, where the expansion occurs, were expressed 10-fold higher in mutation carriers than in controls. This may indicate that mutant RNA is selectively stabilized, Edbauer told Alzforum.
In future work, Edbauer plans to more closely investigate how DPR proteins are expressed, and test each of them for toxicity in cell culture. Based on those studies, he will make a mouse model that accumulates the aberrant proteins. He is also continuing human work to see how closely DPR inclusions correlate with disease, and to discover if the proteins are present in the cerebrospinal fluid and might serve as a biomarker of the disease.
John Trojanowski at the University of Pennsylvania, Philadelphia, called the study “ingenious,” and said that the evidence for DPR translation and accumulation adds an important piece to the ALS/FTLD puzzle. “Having another player to consider in mechanisms of disease is important,” he said. He added that the knowledge may aid researchers in making effective animal models of the disorder.
“I think it is very important work. We had assumed the DENNL72 [C9ORF72] mutation caused disease either through RNA toxicity or through loss of function. This suggests a third and very plausible mechanism: gain of toxic protein function,” John Hardy at University College London, U.K., wrote to Alzforum.
Ranum, now at the University of Florida, Gainesville, was also impressed by the work. “It’s an exciting study because it highlights the possibility that protein gain of function plays a role in diseases that are thought to be RNA gain of function,” she told Alzforum. Both mechanisms may contribute to disease, and could be targeted by killing the messenger RNA, she suggested. “This mechanism of RAN translation may have a role in many expansion diseases. This could be the tip of the iceberg.”—Madolyn Bowman Rogers
References
News Citations
- Corrupt Code: DNA Repeats Are Common Cause for ALS and FTD
- C9ORF72 Steals the Show at Frontotemporal Dementia Meeting
- C9ORF72 Function: Is the ALS Protein a Membrane Traffic Cop?
- Chicago—RNA Inclusions Offer Therapeutic Target in ALS
Paper Citations
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, Nan Z, Forster C, Low WC, Schoser B, Somia NV, Clark HB, Schmechel S, Bitterman PB, Gourdon G, Swanson MS, Moseley M, Ranum LP. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011 Jan 4;108(1):260-5. Epub 2010 Dec 20 PubMed.
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobágyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011 Dec;122(6):691-702. PubMed.
- Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC, Rossor MN, Hardy J, Collinge J, Revesz T, Mead S, Warren JD. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain. 2012 Mar;135(Pt 3):736-50. PubMed.
- Bigio EH, Weintraub S, Rademakers R, Baker M, Ahmadian SS, Rademaker A, Weitner BB, Mao Q, Lee KH, Mishra M, Ganti RA, Mesulam MM. Frontotemporal lobar degeneration with TDP-43 proteinopathy and chromosome 9p repeat expansion in C9ORF72: clinicopathologic correlation. Neuropathology. 2012 Jun 18; PubMed.
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011 Oct 20;72(2):245-56. Epub 2011 Sep 21 PubMed.
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Bäumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M, Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012 Jan;11(1):54-65. PubMed.
Other Citations
Further Reading
News
- C9ORF72 Update: ALS Gene Is a Variable, and Global, Phenomenon
- When Is a C9ORF72 Repeat Expansion Not a C9ORF72 Repeat Expansion?
- Chicago—Dynamic Repeats: C9ORF72 Expands and Shrinks in ALS
- Corrupt Code: DNA Repeats Are Common Cause for ALS and FTD
- C9ORF72 Steals the Show at Frontotemporal Dementia Meeting
- C9ORF72 Function: Is the ALS Protein a Membrane Traffic Cop?
- Chicago—RNA Inclusions Offer Therapeutic Target in ALS
Primary Papers
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013 Mar 15;339(6125):1335-8. Epub 2013 Feb 7 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
The Hospital for Sick Children
This report shows an unusual form of biochemistry that contributes to certain forms of ALS/FTLD. Typically, DNA is transcribed to make an RNA, and the RNA is translated to make a protein. This translation almost always requires a signal that says “Translation begins here.” This signal is an ATG codon in the sequence. Moreover, typically one RNA produces only one protein. Most, but not all, forms of ALS/FTLD are caused by an expanded GGGGCC repeat in the DNA of the C9ORF72 gene which is transcribed to an RNA with the expanded repeat. This paper suggests that this repeat is translated in a manner that does not require that start signal. This repeat-associated non-ATG translation (RAN-translation) of the C9ORF72 hexanucleotide repeat resembles that initially observed by Laura P. Ranum's lab for myotonic dystrophy type 1 and spinocerebellar ataxia type 8 trinucleotide repeats, CTG, and CAG (Zu et al., 2011). For the ALS/FTLD expanded repeat, three different RNA proteins are produced and present in patients' brains. The presence of RAN-translated protein products in brains of ALS/FTLD patients who have the repeat expansion mutation, but not in patients with other variants of ALS/FTLD without the expansion, or in non-affected individuals, is striking for several reasons: First, this finding provides further support for the existence of this totally strange form of translation initially observed by Ranum and colleagues. Second, it reveals the potential involvement of dipeptide repeats in pathogenesis. Third, it further subclassifies the forms of this serious set of motor neuron diseases; ALS/FTLD with and without expansions and RAN-translation products. These are exciting advances for ALS/FTLD research, but only more research will tell what is the role, if any, of these RAN products in disease. Stay tuned!
References:
Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, Nan Z, Forster C, Low WC, Schoser B, Somia NV, Clark HB, Schmechel S, Bitterman PB, Gourdon G, Swanson MS, Moseley M, Ranum LP. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011 Jan 4;108(1):260-5. Epub 2010 Dec 20 PubMed.
Institut Jožef Stefan
This paper represents an important step in understanding the molecular processes in FTLD/ALS, and expands the basic concepts of biological processes. Furthermore, the dipeptide repeat (DPR) protein antibodies will provide a powerful research and diagnostic tool for our field.
The big question now is the relevance of DPRs to disease. The DPR expression is reported in patient cerebellum and hippocampus, which do not have a direct connection with FTLD/ALS; therefore, are DPRs expressed to that level in regions directly connected with the disease, such as frontal cortex or spinal cord? As far as it is known, repeat-associated non-ATG-initiated (RAN) translation requires hairpin formation. Following suggestions that GGGGCC may form G-quadruplex structures (in vitro), several explanations for the translation and its anatomical distribution come to mind: 1) RAN translation can be initiated also from G-quadruplex structures; 2) G-guadruplexes do not form in vivo and the hairpin is the major structure of the RNA repeat; 3) different regions of the brain have a different complement of RNA-binding factors, and hippocampus and cerebellum may have an environment inducive for hairpin formation. Those conditions may be absent in frontotemporal cortex and spinal cord; 4) there are temporal and spatial differences in the expression level of the GGGGCC repeat, such that GGGGCC repeat expression is increased in hippocampus and cerebellum with the onset of disease.
Make a Comment
To make a comment you must login or register.